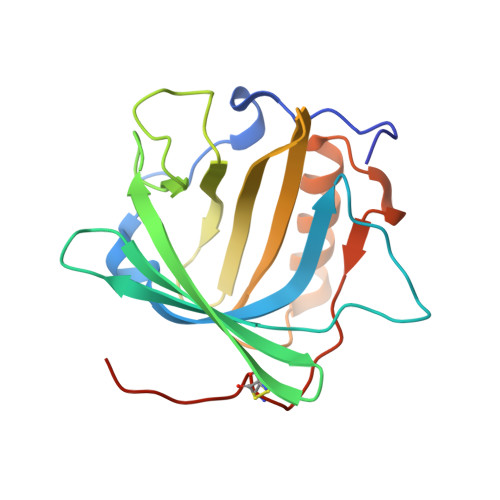
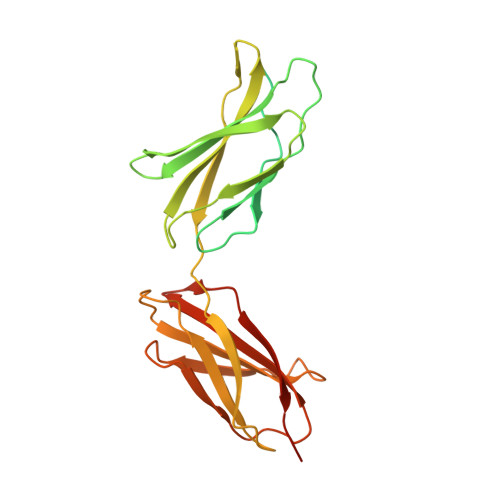
Anticalins Reveal High Plasticity in the Mode of Complex Formation with a Common Tumor Antigen.
Schiefner, A., Gebauer, M., Richter, A., Skerra, A.(2018) Structure 26: 649-656.e3
- PubMed: 29526433
- DOI: https://doi.org/10.1016/j.str.2018.02.003
- Primary Citation of Related Structures:
5N47, 5N48 - PubMed Abstract:
We describe the comparative X-ray structural analysis of three Anticalin proteins directed against the extra-domain B (ED-B) of oncofetal fibronectin (Fn), a validated marker of tumor neoangiogenesis. The Anticalins were engineered from the human lipocalin 2 (Lcn2) scaffold via targeted randomization of the structurally variable loop region and selection by phage display, resulting in 15-19 exchanged residues. While the four reshaped loops exhibit diverse conformations (with shifts in Cα positions up to 20.4 Å), the β-barrel core of the lipocalin remains strongly conserved, thus confirming the extraordinary robustness of this scaffold. All three Anticalins bind the cc' hairpin loop of ED-B, the most exposed motif in the context of its neighboring Fn domains, but reveal entirely different binding modes, with orientations differing by up to 180°. Hence, each Anticalin recognizes its molecular target in an individual manner, in line with the distinct epitope specificities previously seen in binding experiments.
Organizational Affiliation:
Munich Center for Integrated Protein Science (CIPS-M) and Lehrstuhl für Biologische Chemie, Technische Universität München, Emil-Erlenmeyer-Forum 5, 85354 Freising (Weihenstephan), Germany.