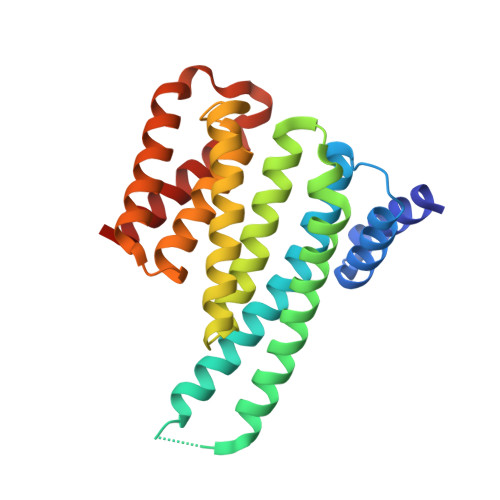
Structural Basis for the Interaction of a Human Small Heat Shock Protein with the 14-3-3 Universal Signaling Regulator.
Sluchanko, N.N., Beelen, S., Kulikova, A.A., Weeks, S.D., Antson, A.A., Gusev, N.B., Strelkov, S.V.(2017) Structure 25: 305-316
- PubMed: 28089448
- DOI: https://doi.org/10.1016/j.str.2016.12.005
- Primary Citation of Related Structures:
5LTW, 5LU1, 5LU2, 5LUM - PubMed Abstract:
By interacting with hundreds of protein partners, 14-3-3 proteins coordinate vital cellular processes. Phosphorylation of the small heat shock protein, HSPB6, within its intrinsically disordered N-terminal domain activates its interaction with 14-3-3, ultimately triggering smooth muscle relaxation. After analyzing the binding of an HSPB6-derived phosphopeptide to 14-3-3 using isothermal calorimetry and X-ray crystallography, we have determined the crystal structure of the complete assembly consisting of the 14-3-3 dimer and full-length HSPB6 dimer and further characterized this complex in solution using fluorescence spectroscopy, small-angle X-ray scattering, and limited proteolysis. We show that selected intrinsically disordered regions of HSPB6 are transformed into well-defined conformations upon the interaction, whereby an unexpectedly asymmetric structure is formed. This structure provides the first atomic resolution snapshot of a human small HSP in functional state, explains how 14-3-3 proteins sequester their regulatory partners, and can inform the design of small-molecule interaction modifiers to be used as myorelaxants.
Organizational Affiliation:
Laboratory of Structural Biochemistry of Proteins, A.N. Bach Institute of Biochemistry, Federal Research Center "Fundamentals of Biotechnology", Russian Academy of Sciences, 119071 Moscow, Russia. Electronic address: nikolai.sluchanko@mail.ru.