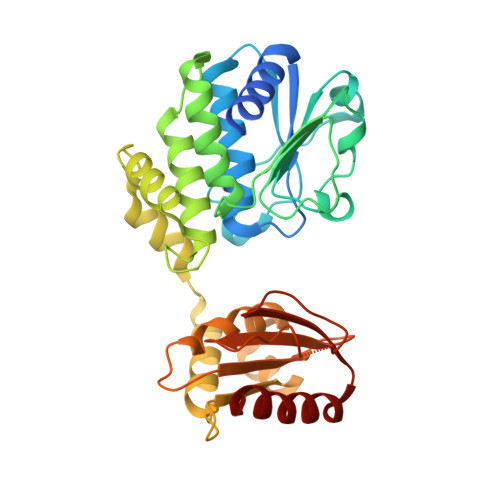
Structural Basis for the Bidirectional Activity of Bacillus nanoRNase NrnA.
Schmier, B.J., Nelersa, C.M., Malhotra, A.(2017) Sci Rep 7: 11085-11085
- PubMed: 28894100
- DOI: https://doi.org/10.1038/s41598-017-09403-x
- Primary Citation of Related Structures:
5IPP, 5IUF, 5IZO, 5J21 - PubMed Abstract:
NanoRNAs are RNA fragments 2 to 5 nucleotides in length that are generated as byproducts of RNA degradation and abortive transcription initiation. Cells have specialized enzymes to degrade nanoRNAs, such as the DHH phosphoesterase family member NanoRNase A (NrnA). This enzyme was originally identified as a 3' → 5' exonuclease, but we show here that NrnA is bidirectional, degrading 2-5 nucleotide long RNA oligomers from the 3' end, and longer RNA substrates from the 5' end. The crystal structure of Bacillus subtilis NrnA reveals a dynamic bi-lobal architecture, with the catalytic N-terminal DHH domain linked to the substrate binding C-terminal DHHA1 domain via an extended linker. Whereas this arrangement is similar to the structure of RecJ, a 5' → 3' DHH family DNase and other DHH family nanoRNases, Bacillus NrnA has gained an extended substrate-binding patch that we posit is responsible for its 3' → 5' activity.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Miami Miller School of Medicine, PO Box 016129, Miami, FL, 33101-6129, USA.