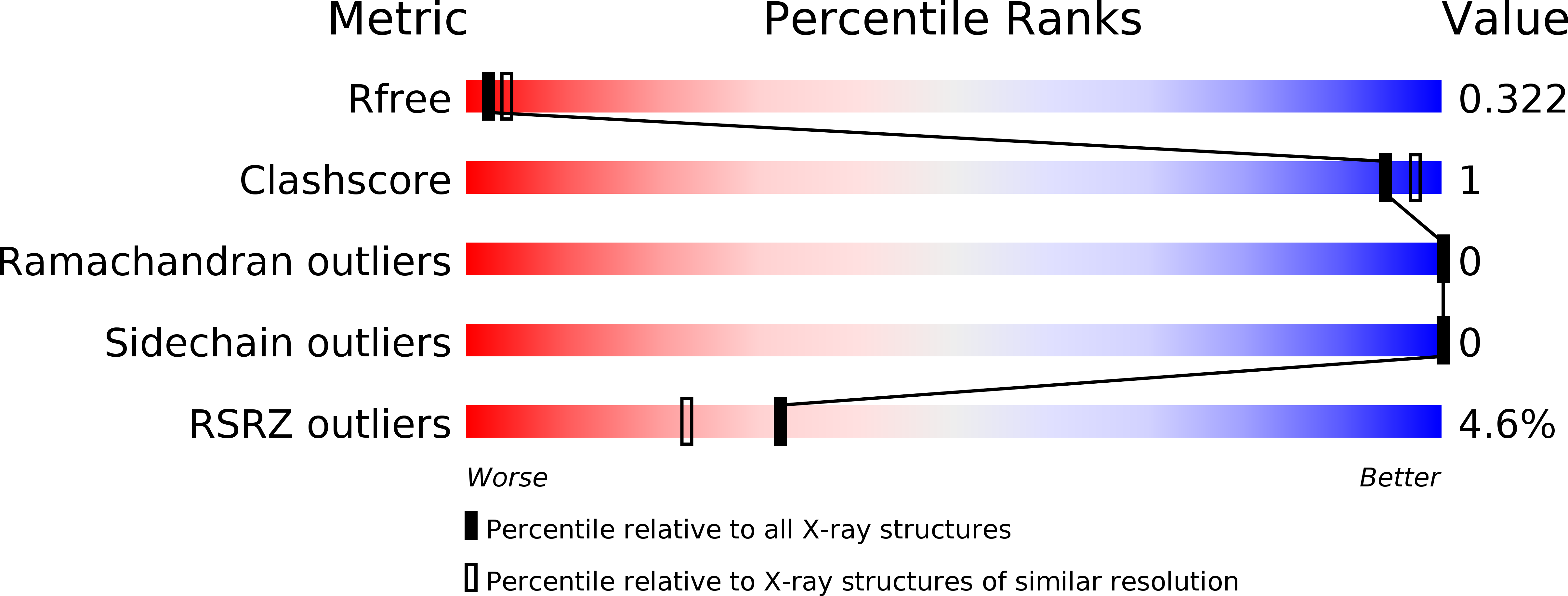
The Structure of the Plakin Domain of Plectin Reveals an Extended Rod-like Shape.
Ortega, E., Manso, J.A., Buey, R.M., Carballido, A.M., Carabias, A., Sonnenberg, A., de Pereda, J.M.(2016) J Biol Chem 291: 18643-18662
- PubMed: 27413182
- DOI: https://doi.org/10.1074/jbc.M116.732909
- Primary Citation of Related Structures:
5J1F, 5J1G, 5J1H, 5J1I - PubMed Abstract:
Plakins are large multi-domain proteins that interconnect cytoskeletal structures. Plectin is a prototypical plakin that tethers intermediate filaments to membrane-associated complexes. Most plakins contain a plakin domain formed by up to nine spectrin repeats (SR1-SR9) and an SH3 domain. The plakin domains of plectin and other plakins harbor binding sites for junctional proteins. We have combined x-ray crystallography with small angle x-ray scattering (SAXS) to elucidate the structure of the plakin domain of plectin, extending our previous analysis of the SR1 to SR5 region. Two crystal structures of the SR5-SR6 region allowed us to characterize its uniquely wide inter-repeat conformational variability. We also report the crystal structures of the SR7-SR8 region, refined to 1.8 Å, and the SR7-SR9 at lower resolution. The SR7-SR9 region, which is conserved in all other plakin domains, forms a rigid segment stabilized by uniquely extensive inter-repeat contacts mediated by unusually long helices in SR8 and SR9. Using SAXS we show that in solution the SR3-SR6 and SR7-SR9 regions are rod-like segments and that SR3-SR9 of plectin has an extended shape with a small central kink. Other plakins, such as bullous pemphigoid antigen 1 and microtubule and actin cross-linking factor 1, are likely to have similar extended plakin domains. In contrast, desmoplakin has a two-segment structure with a central flexible hinge. The continuous versus segmented structures of the plakin domains of plectin and desmoplakin give insight into how different plakins might respond to tension and transmit mechanical signals.
Organizational Affiliation:
From the Instituto de Biología Molecular y Celular del Cancer, Consejo Superior de Investigaciones Científicas, University of Salamanca, 37007 Salamanca, Spain.