Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Giese, C., Eras, J., Kern, A., Scharer, M.A., Capitani, G., Glockshuber, R.(2016) Angew Chem Int Ed Engl 55: 9350-9355
- PubMed: 27351462
- DOI: https://doi.org/10.1002/anie.201603652
- Primary Citation of Related Structures:
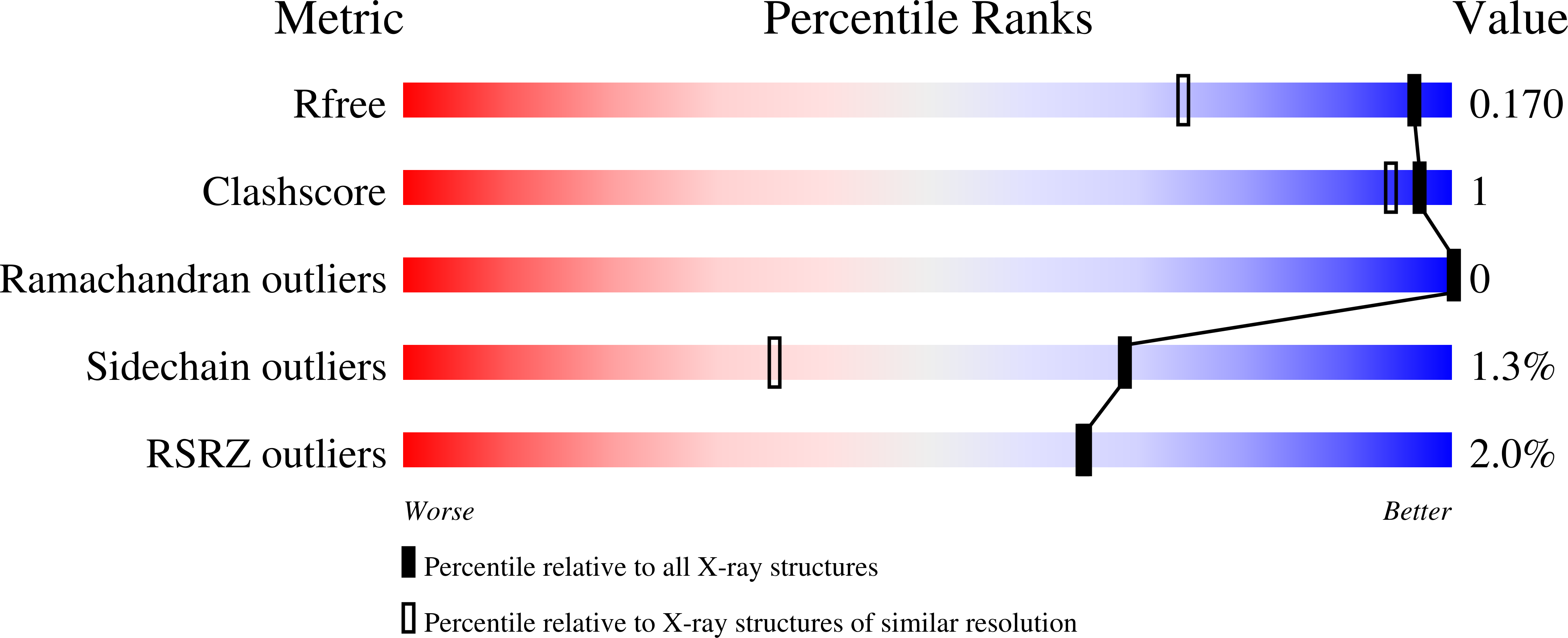
5IQM, 5IQN, 5IQO - PubMed Abstract:
The complex between the bacterial type 1 pilus subunit FimG and the peptide corresponding to the N-terminal extension (termed donor strand, Ds) of the partner subunit FimF (DsF) shows the strongest reported noncovalent molecular interaction, with a dissociation constant (KD ) of 1.5×10(-20) m. However, the complex only exhibits a slow association rate of 330 m(-1) s(-1) that limits technical applications, such as its use in affinity purification. Herein, a structure-based approach was used to design pairs of FimGt (a FimG variant lacking its own N-terminal extension) and DsF variants with enhanced electrostatic surface complementarity. Association of the best mutant FimGt/DsF pairs was accelerated by more than two orders of magnitude, while the dissociation rates and 3D structures of the improved complexes remained essentially unperturbed. A KD value of 8.8×10(-22) m was obtained for the best mutant complex, which is the lowest value reported to date for a protein/ligand complex.
Organizational Affiliation:
Department of Biology, ETH Zurich, Otto-Stern-Weg 5, 8093, Zürich, Switzerland.