Structural Mechanism of Allosteric Activity Regulation in a Ribonucleotide Reductase with Double ATP Cones.
Johansson, R., Jonna, V.R., Kumar, R., Nayeri, N., Lundin, D., Sjoberg, B.M., Hofer, A., Logan, D.T.(2016) Structure 24: 906-917
- PubMed: 27133024
- DOI: https://doi.org/10.1016/j.str.2016.03.025
- Primary Citation of Related Structures:
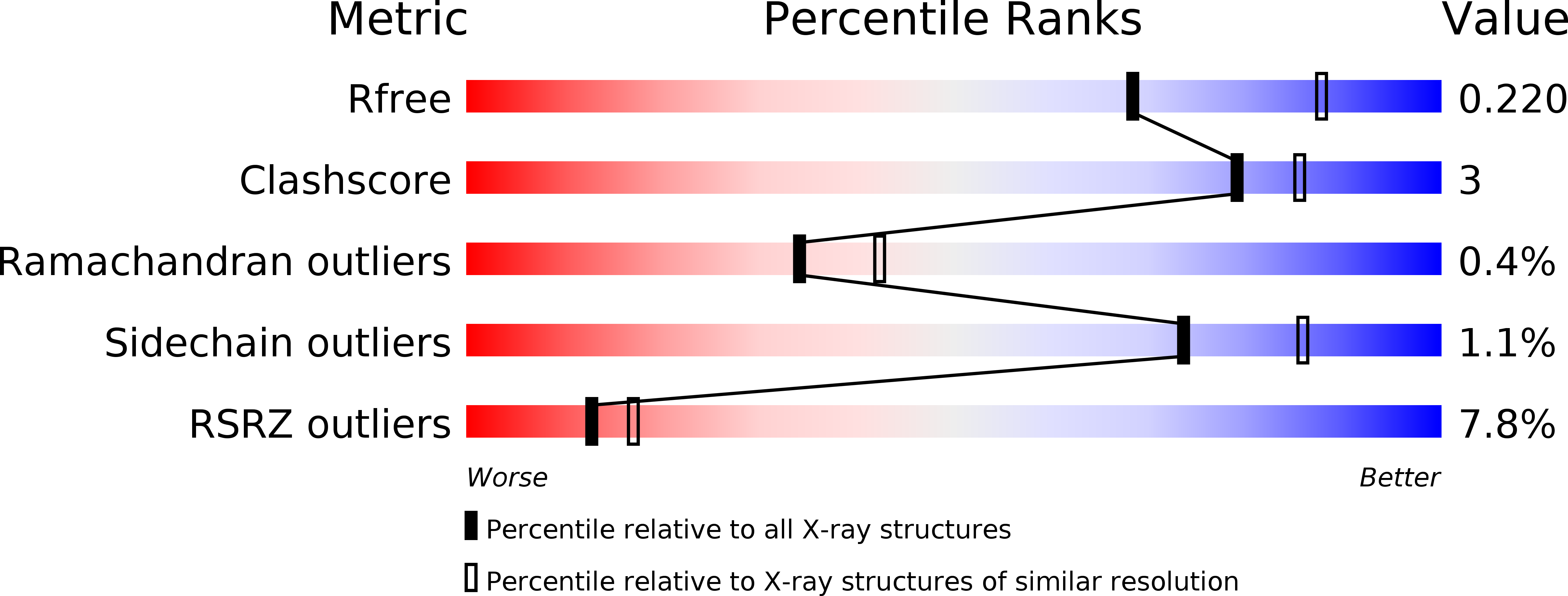
5IM3 - PubMed Abstract:
Ribonucleotide reductases (RNRs) reduce ribonucleotides to deoxyribonucleotides. Their overall activity is stimulated by ATP and downregulated by dATP via a genetically mobile ATP cone domain mediating the formation of oligomeric complexes with varying quaternary structures. The crystal structure and solution X-ray scattering data of a novel dATP-induced homotetramer of the Pseudomonas aeruginosa class I RNR reveal the structural bases for its unique properties, namely one ATP cone that binds two dATP molecules and a second one that is non-functional, binding no nucleotides. Mutations in the observed tetramer interface ablate oligomerization and dATP-induced inhibition but not the ability to bind dATP. Sequence analysis shows that the novel type of ATP cone may be widespread in RNRs. The present study supports a scenario in which diverse mechanisms for allosteric activity regulation are gained and lost through acquisition and evolutionary erosion of different types of ATP cone.
Organizational Affiliation:
Department of Biochemistry & Structural Biology, Lund University, Box 124, 221 00 Lund, Sweden.