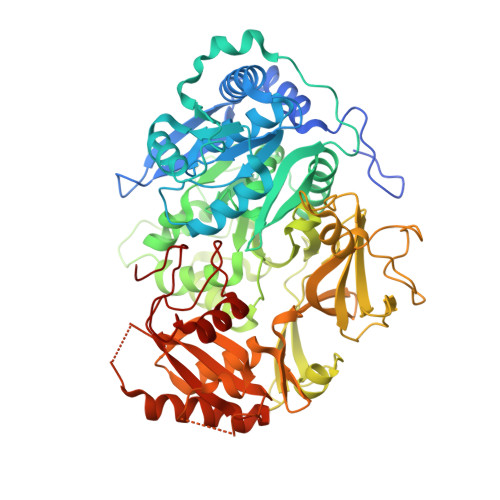
2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.
Minasov, G., Shuvalova, L., Hung, D., Fisher, S.L., Edelstein, J., Kiryukhina, O., Dubrovska, I., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.