Structural and Functional Characterization of a Single-Chain Form of the Recognition Domain of Complement Protein C1q.
Moreau, C., Bally, I., Chouquet, A., Bottazzi, B., Ghebrehiwet, B., Gaboriaud, C., Thielens, N.(2016) Front Immunol 7: 79-79
- PubMed: 26973654
- DOI: https://doi.org/10.3389/fimmu.2016.00079
- Primary Citation of Related Structures:
5HKJ, 5HZF - PubMed Abstract:
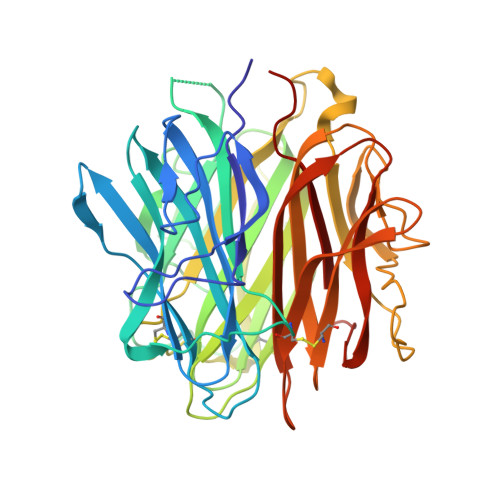
Complement C1q is a soluble pattern recognition molecule comprising six heterotrimeric subunits assembled from three polypeptide chains (A-C). Each heterotrimer forms a collagen-like stem prolonged by a globular recognition domain. These recognition domains sense a wide variety of ligands, including pathogens and altered-self components. Ligand recognition is either direct or mediated by immunoglobulins or pentraxins. Multivalent binding of C1q to its targets triggers immune effector mechanisms mediated via its collagen-like stems. The induced immune response includes activation of the classical complement pathway and enhancement of the phagocytosis of the recognized target. We report here, the first production of a single-chain recombinant form of human C1q globular region (C1q-scGR). The three monomers have been linked in tandem to generate a single continuous polypeptide, based on a strategy previously used for adiponectin, a protein structurally related to C1q. The resulting C1q-scGR protein was produced at high yield in stably transfected 293-F mammalian cells. Recombinant C1q-scGR was correctly folded, as demonstrated by its X-ray crystal structure solved at a resolution of 1.35 Å. Its interaction properties were assessed by surface plasmon resonance analysis using the following physiological C1q ligands: the receptor for C1q globular heads, the long pentraxin PTX3, calreticulin, and heparin. The 3D structure and the binding properties of C1q-scGR were similar to those of the three-chain fragment generated by collagenase digestion of serum-derived C1q. Comparison of the interaction properties of the fragments with those of native C1q provided insights into the avidity component associated with the hexameric assembly of C1q. The interest of this functional recombinant form of the recognition domains of C1q in basic research and its potential biomedical applications are discussed.
Organizational Affiliation:
IBS, University of Grenoble Alpes, Grenoble, France; CNRS, IBS, Grenoble, France; IBS, CEA, Grenoble, France.