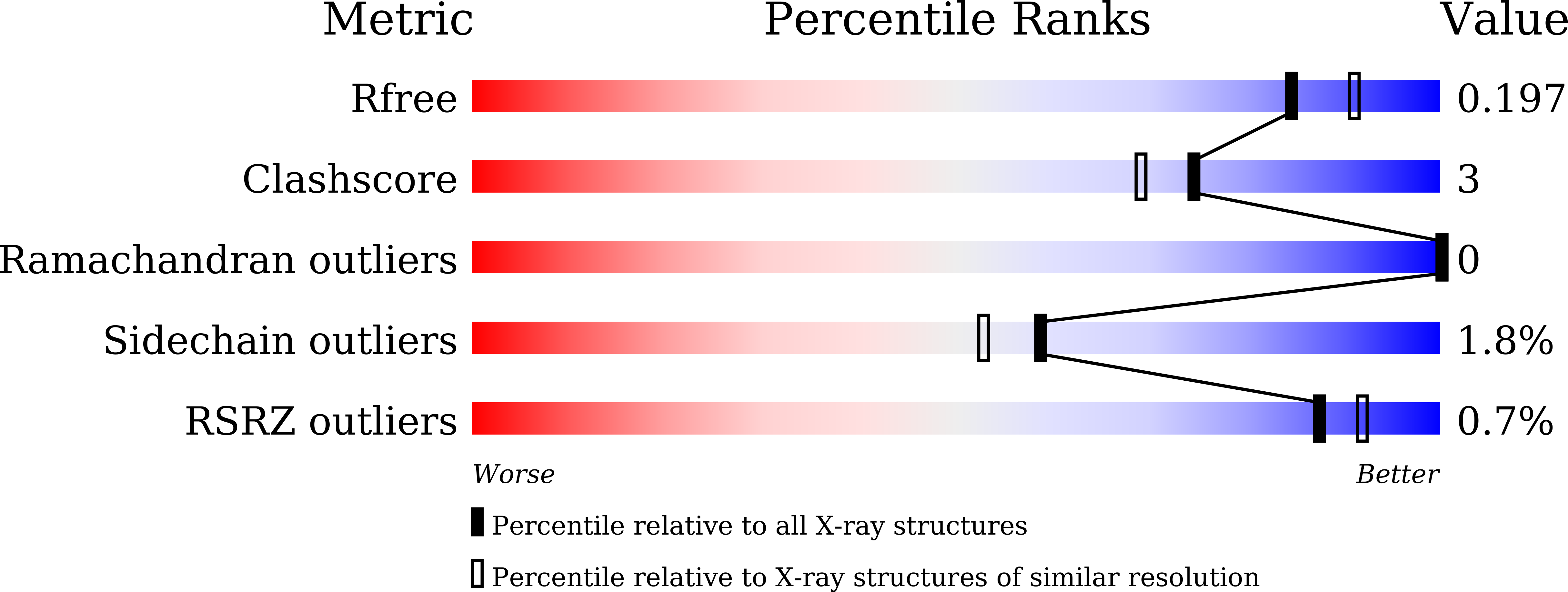
The recognition mechanism of triple-helical beta-1,3-glucan by a beta-1,3-glucanase
Qin, Z., Yang, D., You, X., Liu, Y., Hu, S., Yan, Q., Yang, S., Jiang, Z.(2017) Chem Commun (Camb) 53: 9368-9371
- PubMed: 28787048
- DOI: https://doi.org/10.1039/c7cc03330c
- Primary Citation of Related Structures:
5H9X, 5H9Y - PubMed Abstract:
β-1,3-Glucan is one of the most abundant polysaccharides in fungi. Recognition of β-1,3-glucan occurs in both hydrolysis by glycoside hydrolases and immunological recognition. Our study provides a novel structural account of how glycoside hydrolase recognizes and hydrolyzes substrates in a triple-helical form and presents a general structural basis of β-1,3-glucan recognition.
Organizational Affiliation:
Beijing Advanced Innovation Center for Food Nutrition and Human Health, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing 100083, China. zhqjiang@cau.edu.cn ysq@cau.edu.cn.