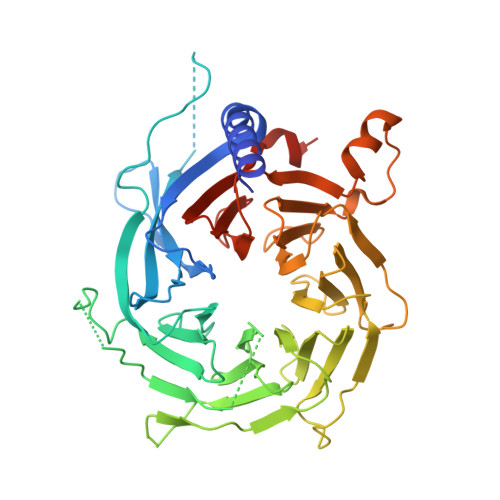
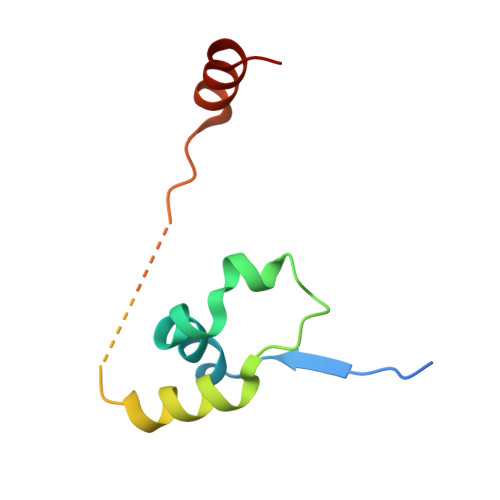
The structure of the core NuRD repression complex provides insights into its interaction with chromatin.
Millard, C.J., Varma, N., Saleh, A., Morris, K., Watson, P.J., Bottrill, A.R., Fairall, L., Smith, C.J., Schwabe, J.W.(2016) Elife 5: e13941-e13941
- PubMed: 27098840
- DOI: https://doi.org/10.7554/eLife.13941
- Primary Citation of Related Structures:
5FXY, 6G16 - PubMed Abstract:
The NuRD complex is a multi-protein transcriptional corepressor that couples histone deacetylase and ATP-dependent chromatin remodelling activities. The complex regulates the higher-order structure of chromatin, and has important roles in the regulation of gene expression, DNA damage repair and cell differentiation. HDACs 1 and 2 are recruited by the MTA1 corepressor to form the catalytic core of the complex. The histone chaperone protein RBBP4, has previously been shown to bind to the carboxy-terminal tail of MTA1. We show that MTA1 recruits a second copy of RBBP4. The crystal structure reveals an extensive interface between MTA1 and RBBP4. An EM structure, supported by SAXS and crosslinking, reveals the architecture of the dimeric HDAC1:MTA1:RBBP4 assembly which forms the core of the NuRD complex. We find evidence that in this complex RBBP4 mediates interaction with histone H3 tails, but not histone H4, suggesting a mechanism for recruitment of the NuRD complex to chromatin.
Organizational Affiliation:
Henry Wellcome Laboratories of Structural Biology, Department of Molecular and Cell Biology, University of Leicester, Leicester, United Kingdom.