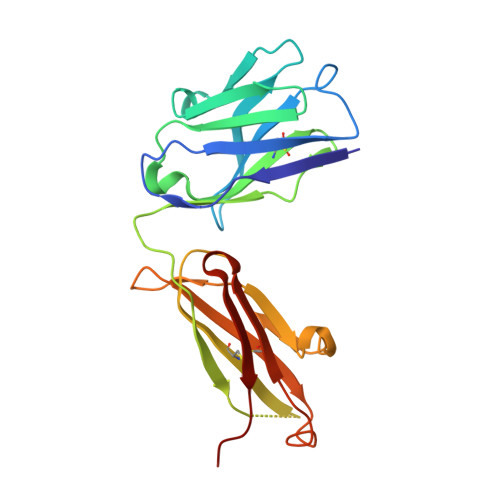
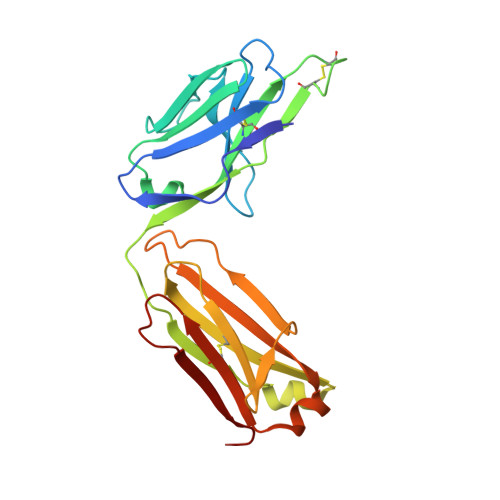
VH-VL orientation prediction for antibody humanization candidate selection: A case study.
Bujotzek, A., Lipsmeier, F., Harris, S.F., Benz, J., Kuglstatter, A., Georges, G.(2016) MAbs 8: 288-305
- PubMed: 26637054
- DOI: https://doi.org/10.1080/19420862.2015.1117720
- Primary Citation of Related Structures:
5DFV, 5DFW, 5DMG - PubMed Abstract:
Antibody humanization describes the procedure of grafting a non-human antibody's complementarity-determining regions, i.e., the variable loop regions that mediate specific interactions with the antigen, onto a β-sheet framework that is representative of the human variable region germline repertoire, thus reducing the number of potentially antigenic epitopes that might trigger an anti-antibody response. The selection criterion for the so-called acceptor frameworks (one for the heavy and one for the light chain variable region) is traditionally based on sequence similarity. Here, we propose a novel approach that selects acceptor frameworks such that the relative orientation of the 2 variable domains in 3D space, and thereby the geometry of the antigen-binding site, is conserved throughout the process of humanization. The methodology relies on a machine learning-based predictor of antibody variable domain orientation that has recently been shown to improve the quality of antibody homology models. Using data from 3 humanization campaigns, we demonstrate that preselecting humanization variants based on the predicted difference in variable domain orientation with regard to the original antibody leads to subsets of variants with a significant improvement in binding affinity.
Organizational Affiliation:
a Roche Pharmaceutical Research and Early Development, Large Molecule Research, Roche Innovation Center Penzberg , Nonnenwald 2, Penzberg , Germany.