Crystal-Structure-Guided Design of Self-Assembling RNA Nanotriangles.
Boerneke, M.A., Dibrov, S.M., Hermann, T.(2016) Angew Chem Int Ed Engl 55: 4097-4100
- PubMed: 26914842
- DOI: https://doi.org/10.1002/anie.201600233
- Primary Citation of Related Structures:
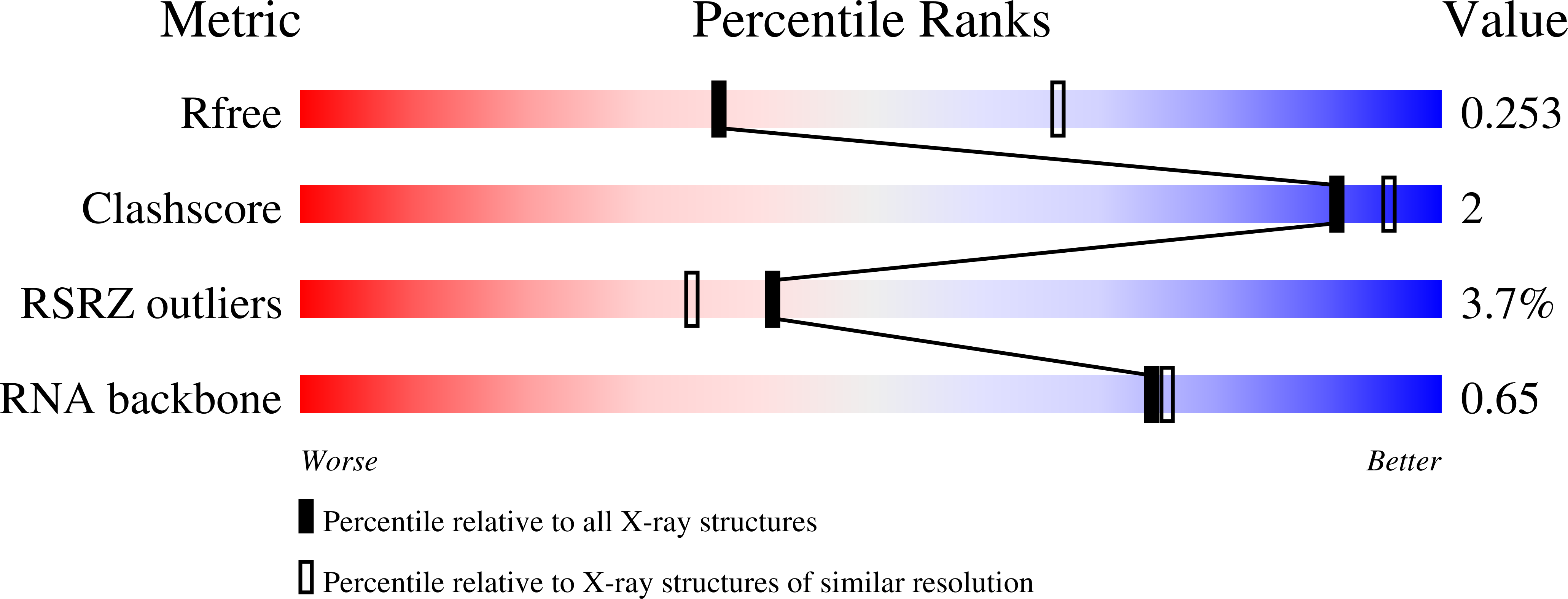
5CNR - PubMed Abstract:
RNA nanotechnology uses RNA structural motifs to build nanosized architectures that assemble through selective base-pair interactions. Herein, we report the crystal-structure-guided design of highly stable RNA nanotriangles that self-assemble cooperatively from short oligonucleotides. The crystal structure of an 81 nucleotide nanotriangle determined at 2.6 Å resolution reveals the so-far smallest circularly closed nanoobject made entirely of double-stranded RNA. The assembly of the nanotriangle architecture involved RNA corner motifs that were derived from ligand-responsive RNA switches, which offer the opportunity to control self-assembly and dissociation.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, CA, 92093, USA.