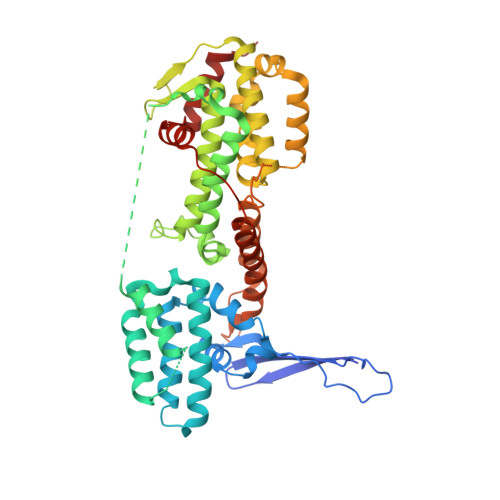
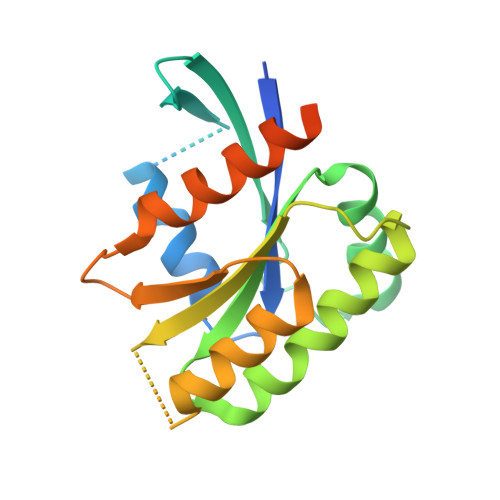
The structure of the Guanine Nucleotide Exchange Factor Rlf in complex with the small G-protein Ral identifies conformational intermediates of the exchange reaction and the basis for the selectivity.
Popovic, M., Schouten, A., Rensen-de Leeuw, M., Rehmann, H.(2016) J Struct Biol 193: 106-114
- PubMed: 26687416
- DOI: https://doi.org/10.1016/j.jsb.2015.12.006
- Primary Citation of Related Structures:
5CM8, 5CM9 - PubMed Abstract:
CDC25 homology domain (CDC25-HD) containing Guanine Nucleotide Exchange Factors (GEFs) initiate signalling by small G-proteins of the Ras-family. Each GEF acts on a small subset of the G-proteins only, thus providing signalling selectivity. Rlf is a GEF with selectivity for the G-proteins RalA and RalB. Here the crystal structure of Rlf in complex with Ral is determined. The Rlf·Ral complex crystallised into two different crystal forms, which represent different steps of the exchange reaction. Thereby general insight in the CDC25-HD catalysed nucleotide exchange is obtained. In addition, the basis for the selectivity of the interaction is investigated. The exchange activity is monitored by the use of recombinant proteins. Selectivity determinants in the binding interface are identified and confirmed by a mutational study.
Organizational Affiliation:
Department of Molecular Cancer Research, Centre of Biomedical Genetics and Cancer Genomics Centre, University Medical Center Utrecht, Utrecht, The Netherlands.