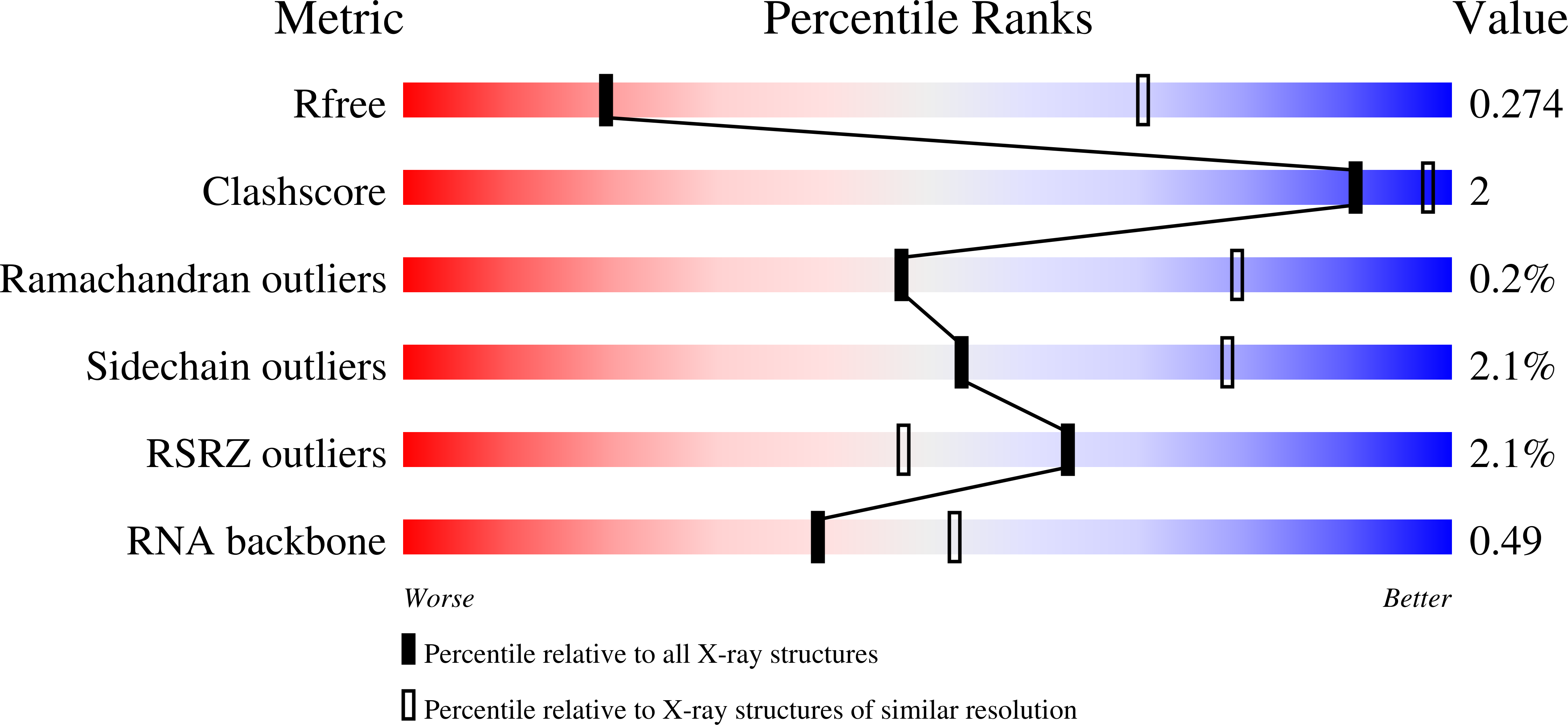
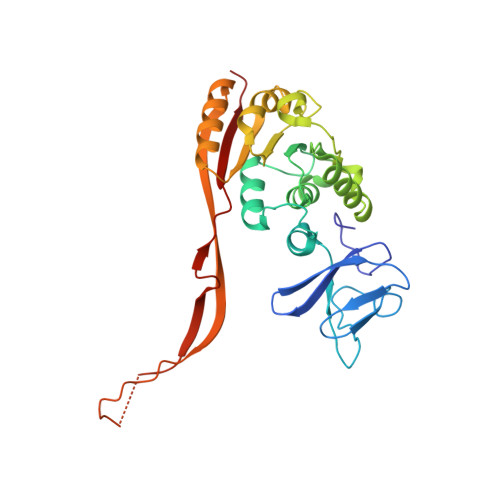
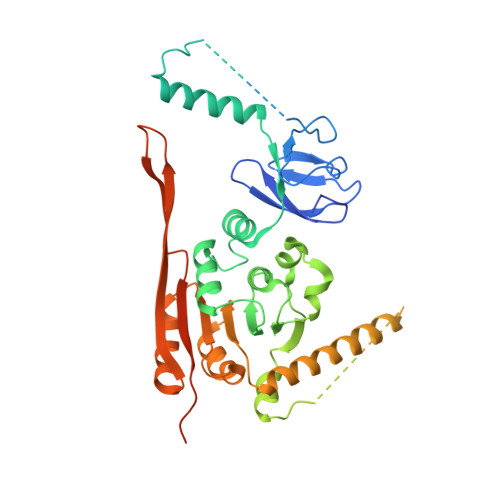
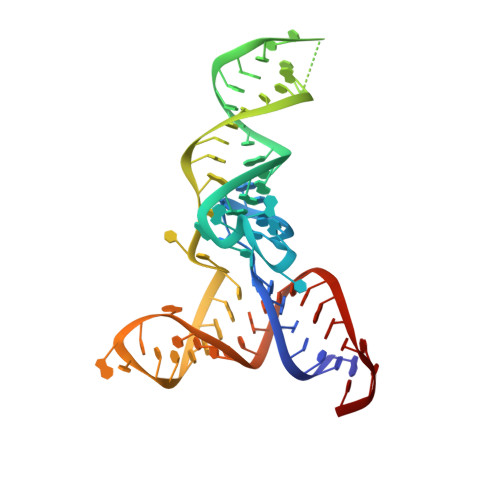
Crystal Structure of the Human tRNA m(1)A58 Methyltransferase-tRNA3(Lys) Complex: Refolding of Substrate tRNA Allows Access to the Methylation Target.
Finer-Moore, J., Czudnochowski, N., O'Connell, J.D., Wang, A.L., Stroud, R.M.(2015) J Mol Biol 427: 3862-3876
- PubMed: 26470919
- DOI: https://doi.org/10.1016/j.jmb.2015.10.005
- Primary Citation of Related Structures:
5CCB, 5CCX, 5CD1 - PubMed Abstract:
Human tRNA3(Lys) is the primer for reverse transcription of HIV; the 3' end is complementary to the primer-binding site on HIV RNA. The complementarity ends at the 18th base, A58, which in tRNA3(Lys) is modified to remove Watson-Crick pairing. Motivated to test the role of the modification in terminating the primer-binding sequence and thus limiting run-on transcription, we asked how the modification of RNA could be accomplished. tRNA m(1)A58 methyltransferase (m(1)A58 MTase) methylates N1 of A58, which is buried in the TΨC-loop of tRNA, from cofactor S-adenosyl-L-methionine. This conserved tRNA modification is essential for stability of initiator tRNA in Saccharomyces cerevisiae. Reported here, three structures of human tRNA m(1)A58 MTase in complex with human tRNA3(Lys) and the product S-adenosyl-L-homocysteine show a dimer of heterodimers in which each heterodimer comprises a catalytic chain, Trm61, and a homologous but noncatalytic chain, Trm6, repurposed as a tRNA-binding subunit that acts in trans; tRNAs bind across the dimer interface such that Trm6 from the opposing heterodimer brings A58 into the active site of Trm61. T-loop and D-loop are splayed apart showing how A58, normally buried in tRNA, becomes accessible for modification. This result has broad impact on our understanding of the mechanisms of modifying internal sites in folded tRNA. The structures serve as templates for design of inhibitors that could be used to test tRNA m(1)A58 MTase's impact on retroviral priming and transcription.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of California San Francisco, San Francisco, CA 94143, USA. Electronic address: finer@msg.ucsf.edu.