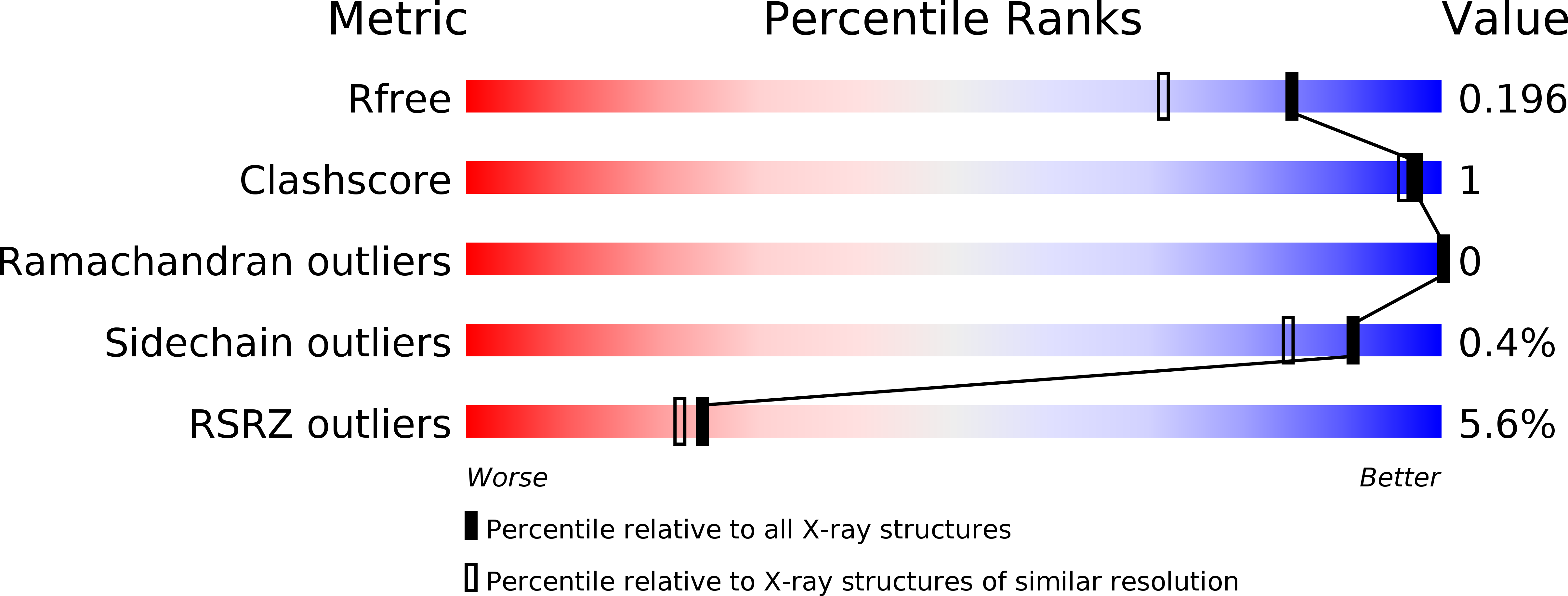
Treatment of norovirus particles with citrate.
Koromyslova, A.D., White, P.A., Hansman, G.S.(2015) Virology 485: 199-204
- PubMed: 26295280
- DOI: https://doi.org/10.1016/j.virol.2015.07.009
- Primary Citation of Related Structures:
5BSX, 5BSY - PubMed Abstract:
Human norovirus is a dominant cause of acute gastroenteritis around the world. Several norovirus disinfectants label citric acid as an active ingredient. In this study, we showed that norovirus virus-like particles (VLPs) treated with citrate buffer caused the particles to alter their morphology, including increased diameters associated with a new ring-like structure. We also found that epitopes on the protruding (P) domain on these particles were more readily accessible to antibodies after the citrate treatment. These results suggested that citrate had a direct effect on the norovirus particles. Using X-ray crystallography, we showed that the P domain bound citrate from lemon juice and a disinfectant containing citric acid. Importantly, citrate binds at the histo-blood group antigen binding pocket, which are attachment factors for norovirus infections. Taken together, these new findings suggested that it might be possible to treat/reduce norovirus infections with citrate, although further studies are needed.
Organizational Affiliation:
Schaller Research Group at the University of Heidelberg and the DKFZ, Heidelberg 69120, Germany; Department of Infectious Diseases, Virology, University of Heidelberg, Heidelberg 69120, Germany.