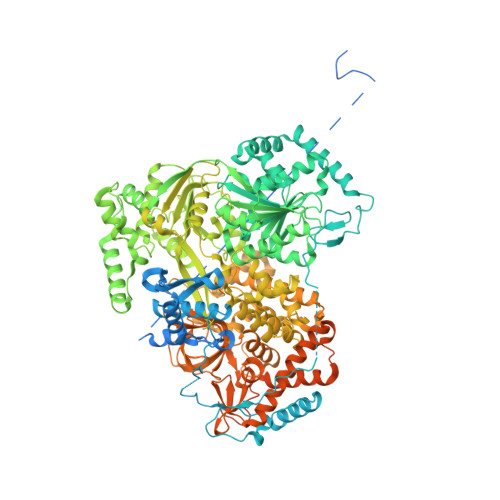
Structure of the RNA Helicase Mle Reveals the Molecular Mechanisms for Uridine Specificity and RNA-ATP Coupling.
Prabu, J.R., Muller, M., Thomae, A.W., Schussler, S., Bonneau, F., Becker, P.B., Conti, E.(2015) Mol Cell 60: 487
- PubMed: 26545078
- DOI: https://doi.org/10.1016/j.molcel.2015.10.011
- Primary Citation of Related Structures:
5AOR - PubMed Abstract:
The MLE helicase remodels the roX lncRNAs, enabling the lncRNA-mediated assembly of the Drosophila dosage compensation complex. We identified a stable MLE core comprising the DExH helicase module and two auxiliary domains: a dsRBD and an OB-like fold. MLEcore is an unusual DExH helicase that can unwind blunt-ended RNA duplexes and has specificity for uridine nucleotides. We determined the 2.1 Å resolution structure of MLEcore bound to a U10 RNA and ADP-AlF4. The OB-like and dsRBD folds bind the DExH module and contribute to form the entrance of the helicase channel. Four uridine nucleotides engage in base-specific interactions, rationalizing the conservation of uridine-rich sequences in critical roX substrates. roX2 binding is orchestrated by MLE's auxiliary domains, which is prerequisite for MLE localization to the male X chromosome. The structure visualizes a transition-state mimic of the reaction and suggests how eukaryotic DEAH/RHA helicases couple ATP hydrolysis to RNA translocation.
Organizational Affiliation:
Department of Structural Cell Biology, Max Planck Institute of Biochemistry, 82152 Martinsried, Germany.