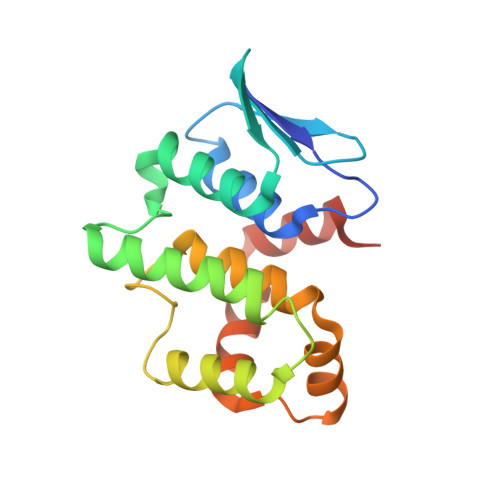
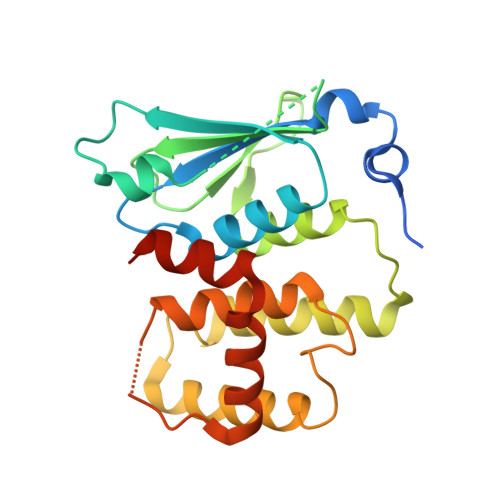
Symmetric Assembly of a Decameric Subcomplex in Human Multi-tRNA Synthetase Complex Via Interactions between Glutathione Transferase-Homology Domains and Aspartyl-tRNA Synthetase.
Cho, H.Y., Lee, H.J., Choi, Y.S., Kim, D.K., Jin, K.S., Kim, S., Kang, B.S.(2019) J Mol Biol
- PubMed: 31473157
- DOI: https://doi.org/10.1016/j.jmb.2019.08.013
- Primary Citation of Related Structures:
5A1N - PubMed Abstract:
Aminoacyl-tRNA synthetases (AARSs) ligate amino acids to their cognate tRNAs during protein synthesis. In humans, eight AARSs and three non-enzymatic AARS-interacting multifunctional proteins (AIMP1-3), which are involved in various biological processes, form a multi-tRNA synthetase complex (MSC). Elucidation of the structures and multiple functions of individual AARSs and AIMPs has aided current understanding of the structural arrangement of MSC components and their assembly processes. Here, we report the crystal structure of a complex comprising a motif from aspartyl-tRNA synthetase (DRS) and the glutathione transferase (GST)-homology domains of methionyl-tRNA synthetase (MRS), glutamyl-prolyl-tRNA synthetase (EPRS), AIMP2, and AIMP3. In the crystal structure, the four GST domains are assembled in the order of MRS-AIMP3-EPRS-AIMP2, and the GST domain of AIMP2 binds DRS through the β-sheet in the GST domain. The C-terminus of AIMP3 enhances the binding of DRS to the tetrameric GST complex. A DRS dimer and two GST tetramers binding to the dimer with 2-fold symmetry complete a decameric complex. The formation of this complex enhances the stability of DRS and enables it to retain its reaction intermediate, aspartyl adenylate. Since the catalytic domains of MRS and EPRS are connected to the decameric complex through their flexible linker peptides, and lysyl-tRNA synthetase and AIMP1 are also linked to the complex via the N-terminal region of AIMP2, the DRS-GST tetramer complex functions as a frame in the MSC.
Organizational Affiliation:
School of Life Science and Biotechnology, KNU Creative BioResearch Group, Kyungpook National University, Daegu 41566, Republic of Korea.