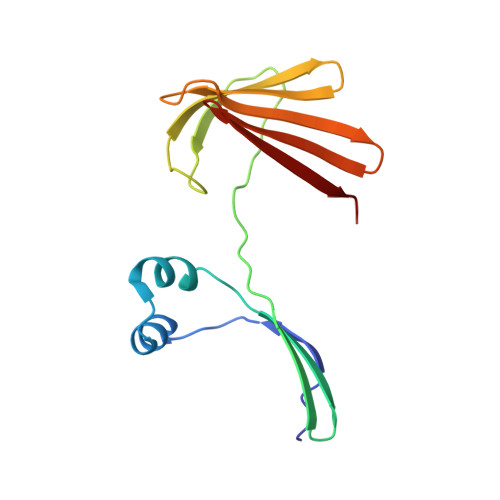
Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Assar, Z., Nossoni, Z., Wang, W., Santos, E.M., Kramer, K., McCornack, C., Vasileiou, C., Borhan, B., Geiger, J.H.(2016) Structure 24: 1590-1598
- PubMed: 27524203
- DOI: https://doi.org/10.1016/j.str.2016.05.022
- Primary Citation of Related Structures:
4ZCB, 4ZGU, 4ZH6, 4ZH9, 4ZJ0, 4ZR2, 5DG4, 5DPQ - PubMed Abstract:
Human Cellular Retinol Binding Protein II (hCRBPII), a member of the intracellular lipid-binding protein family, is a monomeric protein responsible for the intracellular transport of retinol and retinal. Herein we report that hCRBPII forms an extensive domain-swapped dimer during bacterial expression. The domain-swapped region encompasses almost half of the protein. The dimer represents a novel structural architecture with the mouths of the two binding cavities facing each other, producing a new binding cavity that spans the length of the protein complex. Although wild-type hCRBPII forms the dimer, the propensity for dimerization can be substantially increased via mutation at Tyr60. The monomeric form of the wild-type protein represents the thermodynamically more stable species, making the domain-swapped dimer a kinetically trapped entity. Hypothetically, the wild-type protein has evolved to minimize dimerization of the folding intermediate through a critical hydrogen bond (Tyr60-Glu72) that disfavors the dimeric form.
Organizational Affiliation:
Department of Chemistry, Michigan State University, East Lansing, MI 48824, USA.