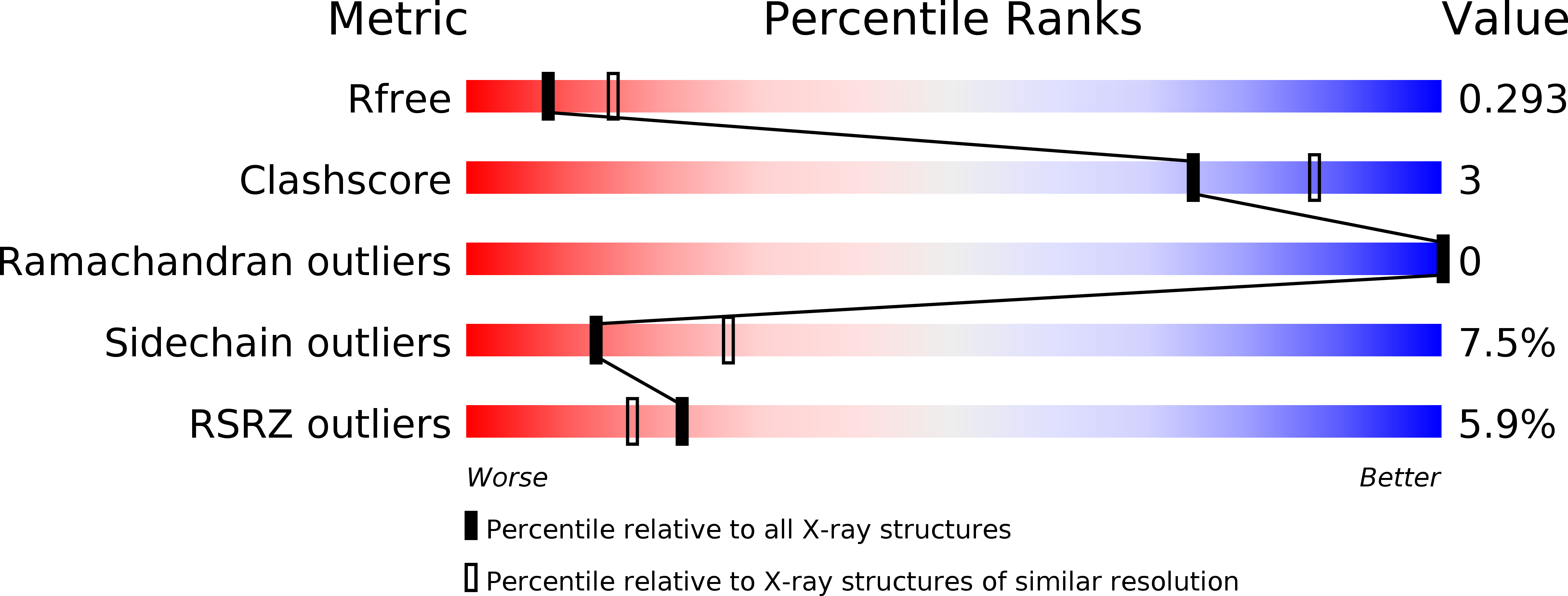
STIL binding to Polo-box 3 of PLK4 regulates centriole duplication.
Arquint, C., Gabryjonczyk, A.M., Imseng, S., Bohm, R., Sauer, E., Hiller, S., Nigg, E.A., Maier, T.(2015) Elife 4
- PubMed: 26188084
- DOI: https://doi.org/10.7554/eLife.07888
- Primary Citation of Related Structures:
2N19, 4YYP - PubMed Abstract:
Polo-like kinases (PLK) are eukaryotic regulators of cell cycle progression, mitosis and cytokinesis; PLK4 is a master regulator of centriole duplication. Here, we demonstrate that the SCL/TAL1 interrupting locus (STIL) protein interacts via its coiled-coil region (STIL-CC) with PLK4 in vivo. STIL-CC is the first identified interaction partner of Polo-box 3 (PB3) of PLK4 and also uses a secondary interaction site in the PLK4 L1 region. Structure determination of free PLK4-PB3 and its STIL-CC complex via NMR and crystallography reveals a novel mode of Polo-box-peptide interaction mimicking coiled-coil formation. In vivo analysis of structure-guided STIL mutants reveals distinct binding modes to PLK4-PB3 and L1, as well as interplay of STIL oligomerization with PLK4 binding. We suggest that the STIL-CC/PLK4 interaction mediates PLK4 activation as well as stabilization of centriolar PLK4 and plays a key role in centriole duplication.
Organizational Affiliation:
Biozentrum, University of Basel, Basel, Switzerland.