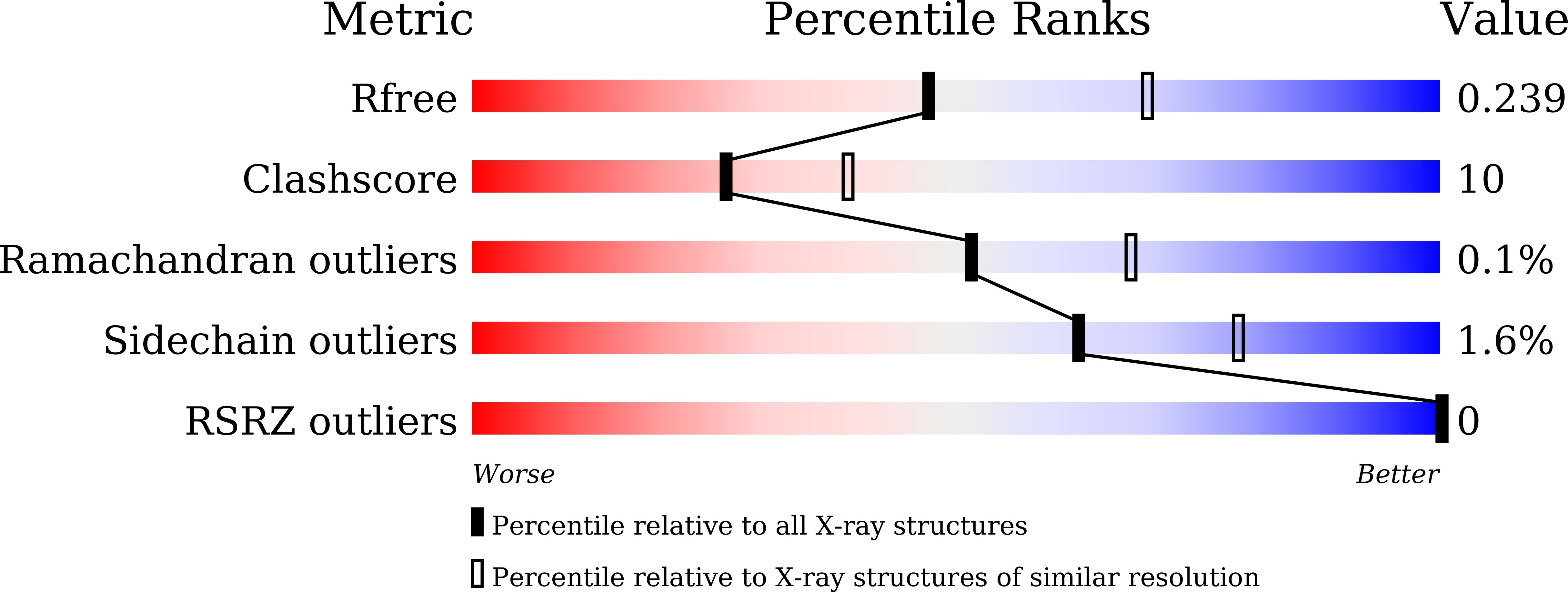
Structural insight into the substrate inhibition mechanism of NADP(+)-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes.
Jang, E.H., Park, S.A., Chi, Y.M., Lee, K.S.(2015) Biochem Biophys Res Commun 461: 487-493
- PubMed: 25888791
- DOI: https://doi.org/10.1016/j.bbrc.2015.04.047
- Primary Citation of Related Structures:
4YWU, 4YWV - PubMed Abstract:
Succinic semialdehyde dehydrogenases (SSADHs) are ubiquitous enzymes that catalyze the oxidation of succinic semialdehyde (SSA) to succinic acid in the presence of NAD(P)(+), and play an important role in the cellular mechanisms including the detoxification of accumulated SSA or the survival in conditions of limited nutrients. Here, we report the inhibitory properties and two crystal structures of SSADH from Streptococcus pyogenes (SpSSADH) in a binary (ES) complex with SSA as the substrate and a ternary (ESS) complex with the substrate SSA and the inhibitory SSA, at 2.4 Å resolution for both structures. Analysis of the kinetic inhibitory parameters revealed significant substrate inhibition in the presence of NADP(+) at concentrations of SSA higher than 0.02 mM, which exhibited complete uncompetitive substrate inhibition with the inhibition constant (Ki) value of 0.10 ± 0.02 mM. In ES-complex of SpSSADH, the SSA showed a tightly bound bent form nearby the catalytic residues, which may be caused by reduction of the cavity volume for substrate binding, compared with other SSADHs. Moreover, structural comparison of ESS-complex with a binary complex with NADP(+) of SpSSADH indicated that the substrate inhibition was induced by the binding of inhibitory SSA in the cofactor-binding site, instead of NADP(+). Our results provide first structure-based molecular insights into the substrate inhibition mechanism of SpSSADH as the Gram-positive bacterial SSADH.
Organizational Affiliation:
Department of Biosystems and Biotechnology, College of Life Sciences and Biotechnology, Korea University, Seoul 136-713, Republic of Korea.