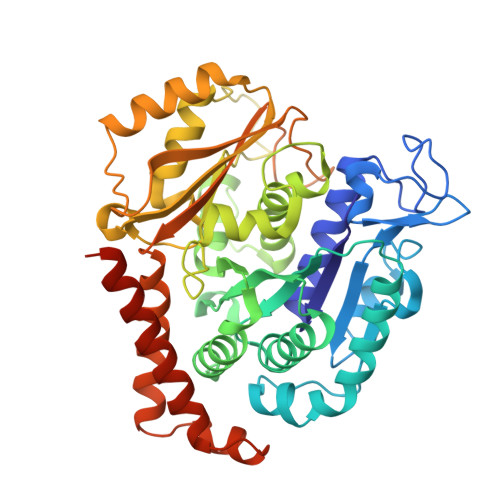
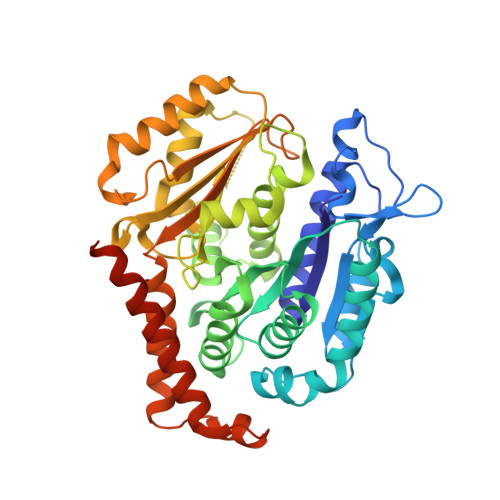
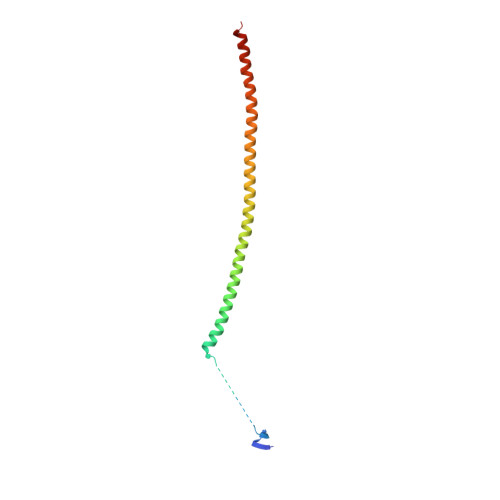
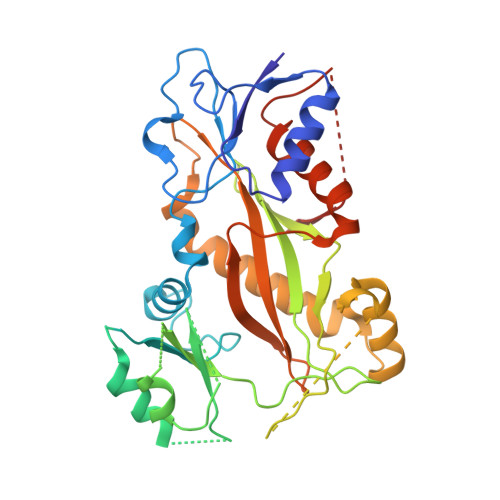
Structures of potent anticancer compounds bound to tubulin.
McNamara, D.E., Senese, S., Yeates, T.O., Torres, J.Z.(2015) Protein Sci 24: 1164-1172
- PubMed: 25970265
- DOI: https://doi.org/10.1002/pro.2704
- Primary Citation of Related Structures:
4YJ2, 4YJ3 - PubMed Abstract:
Small molecules that bind to tubulin exert powerful effects on cell division and apoptosis (programmed cell death). Cell-based high-throughput screening combined with chemo/bioinformatic and biochemical analyses recently revealed a novel compound MI-181 as a potent mitotic inhibitor with heightened activity towards melanomas. MI-181 causes tubulin depolymerization, activates the spindle assembly checkpoint arresting cells in mitosis, and induces apoptotic cell death. C2 is an unrelated compound previously shown to have lethal effects on microtubules in tumorigenic cell lines. We report 2.60 Å and 3.75 Å resolution structures of MI-181 and C2, respectively, bound to a ternary complex of αβ-tubulin, the tubulin-binding protein stathmin, and tubulin tyrosine ligase. In the first of these structures, our crystallographic results reveal a unique binding mode for MI-181 extending unusually deep into the well-studied colchicine-binding site on β-tubulin. In the second structure the C2 compound occupies the colchicine-binding site on β-tubulin with two chemical moieties recapitulating contacts made by colchicine, in combination with another system of atomic contacts. These insights reveal the source of the observed effects of MI-181 and C2 on microtubules, mitosis, and cultured cancer cell lines. The structural details of the interaction between tubulin and the described compounds may guide the development of improved derivative compounds as therapeutic candidates or molecular probes to study cancer cell division.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Los Angeles, Los Angeles, California, 90095.