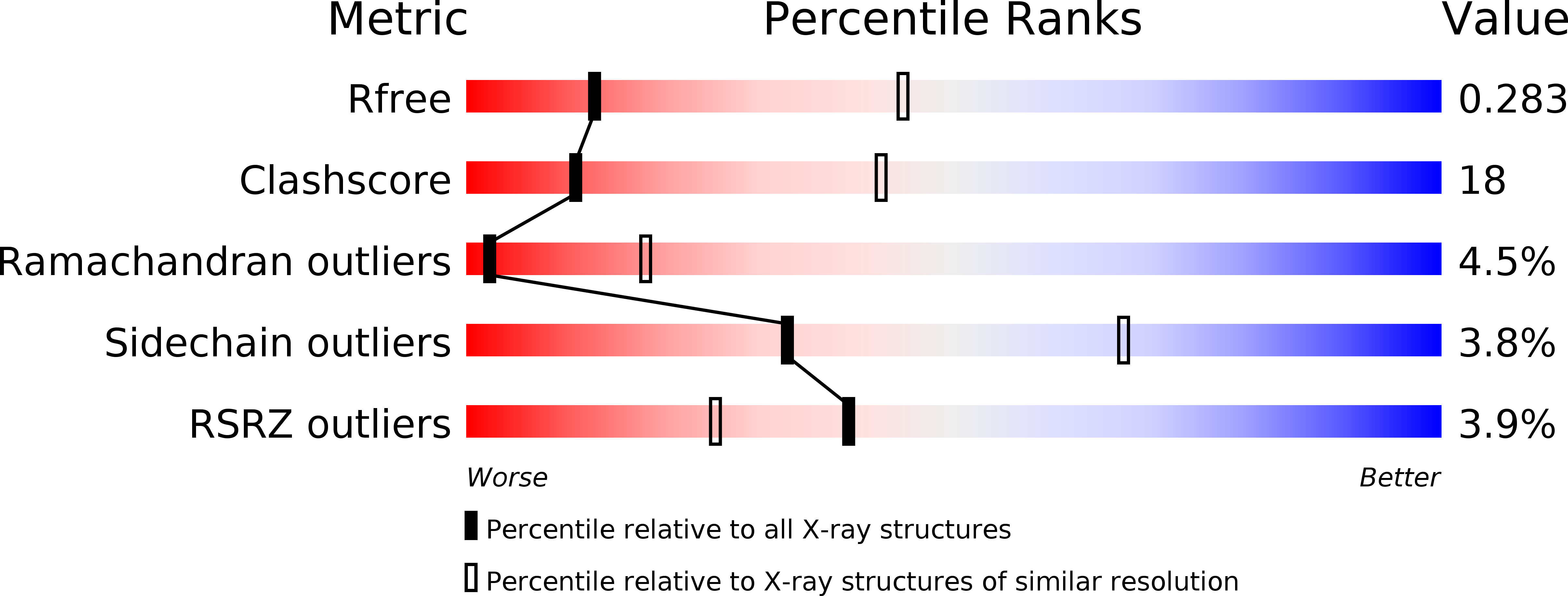
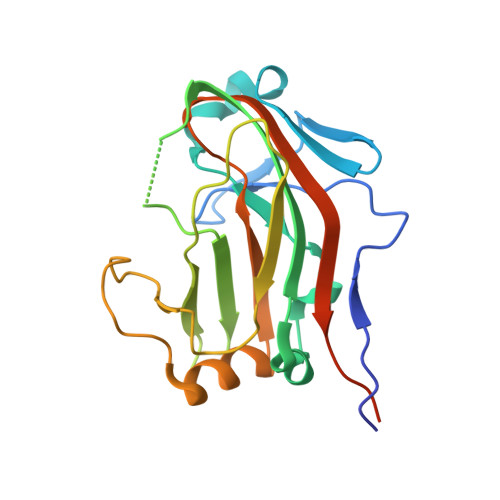
Laminin L4 domain structure resembles adhesion modules in ephrin receptor and other transmembrane glycoproteins.
Moran, T., Gat, Y., Fass, D.(2015) FEBS J 282: 2746-2757
- PubMed: 25962468
- DOI: https://doi.org/10.1111/febs.13319
- Primary Citation of Related Structures:
4YEP, 4YEQ - PubMed Abstract:
The ~ 800 kDa laminin heterotrimer forms a distinctive cross-shaped structure that further self-assembles into networks within the extracellular matrix. The domains at the laminin chain termini, which engage in network formation and cell-surface interaction, are well understood both structurally and functionally. By contrast, the structures and roles of additional domains embedded within the limbs of the laminin cross have remained obscure. Here, we report the X-ray crystal structure, determined to 1.2 Å resolution, of the human laminin α2 subunit L4b domain, site of an inframe deletion mutation associated with mild congenital muscular dystrophy. The α2 L4b domain is an irregular β-sandwich with many short and broken strands linked by extended loops. The most similar known structures are the carbohydrate-binding domains of bacterial cellulases, the ephrin-binding domain of ephrin receptors, and MAM adhesion domains in various other eukaryotic cell-surface proteins. This similarity to mammalian adhesion modules, which was not predicted on the basis of amino acid sequence alone due to lack of detectable homology, suggests that laminin internal domains evolved from a progenitor adhesion molecule and may retain a role in cell adhesion in the context of the laminin trimer. The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ, USA (http://www.rcsb.org/) under codes 4YEP and 4YEQ.
Organizational Affiliation:
Department of Structural Biology, Weizmann Institute of Science, Rehovot, Israel.