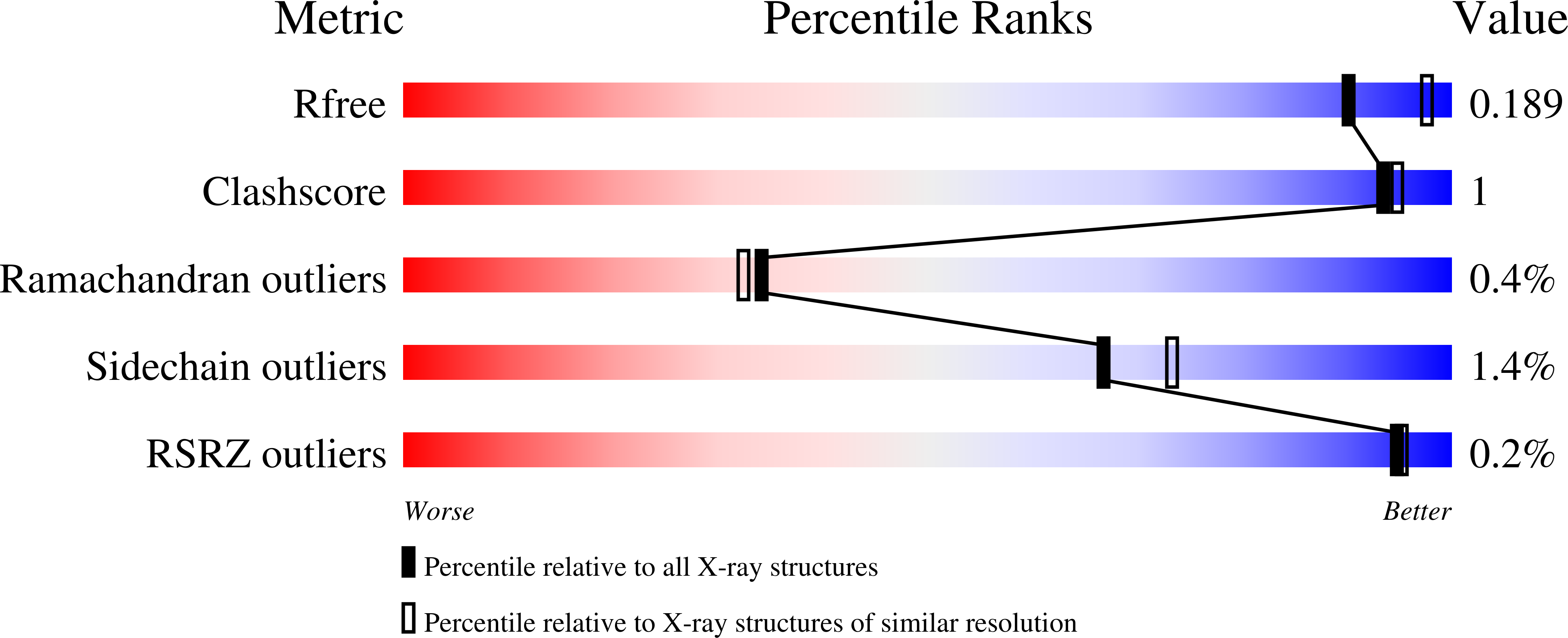
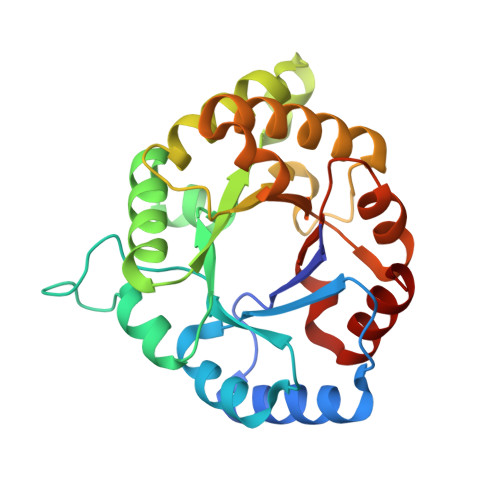
Reversibility and two state behaviour in the thermal unfolding of oligomeric TIM barrel proteins.
Romero-Romero, S., Costas, M., Rodriguez-Romero, A., Alejandro Fernandez-Velasco, D.(2015) Phys Chem Chem Phys 17: 20699-20714
- PubMed: 26206330
- DOI: https://doi.org/10.1039/c5cp01599e
- Primary Citation of Related Structures:
4Y8F, 4Y90, 4Y96, 4Y9A - PubMed Abstract:
Temperature is one of the main variables that modulate protein function and stability. Thermodynamic studies of oligomeric proteins, the dominant protein natural form, have been often hampered because irreversible aggregation and/or slow reactions are common. There are no reports on the reversible equilibrium thermal unfolding of proteins composed of (β/α)8 barrel subunits, albeit this "TIM barrel" topology is one of the most abundant and versatile in nature. We studied the eponymous TIM barrel, triosephosphate isomerase (TIM), belonging to five species of different bacterial taxa. All of them were found to be catalytically efficient dimers. The three-dimensional structure of four enzymes was solved at high/medium resolution. Irreversibility and kinetic control were observed in the thermal unfolding of two TIMs, while for the other three the thermal unfolding was found to follow a two-state equilibrium reversible process. Shifts in the global stability curves of these three proteins are related to the organismal temperature range of optimal growth and modulated by variations in maximum stability temperature and in the enthalpy change at that temperature. Reversibility appears to correlate with the low isoelectric point, the absence of a residual structure in the unfolded state, small cavity volume in the native state, low conformational stability and a low melting temperature. Furthermore, the strong coupling between dimer dissociation and monomer unfolding may reduce aggregation and favour reversibility. It is therefore very thought-provoking to find that a common topological ensemble, such as the TIM barrel, can unfold/refold in the Anfinsen way, i.e. without the help of the cellular machinery.
Organizational Affiliation:
Laboratorio de Fisicoquímica e Ingeniería de Proteínas, Departamento de Bioquímica, Facultad de Medicina, Universidad Nacional Autónoma de México, 04510 Ciudad de México, Distrito Federal, Mexico. fdaniel@unam.mx.