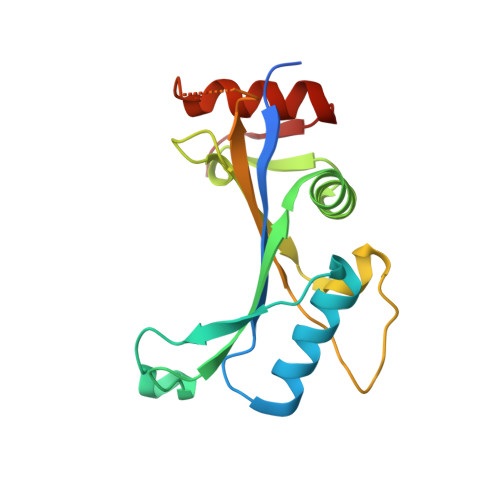
Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation
Zheng, J., Lang, Y., Zhang, Q., Cui, D., Sun, H., Jiang, L., Chen, Z., Zhang, R., Gao, Y., Tian, W., Wu, W., Tang, J., Chen, Z.(2015) Genes Dev 29: 1524-1534
- PubMed: 26220995
- DOI: https://doi.org/10.1101/gad.261792.115
- Primary Citation of Related Structures:
4XXB - PubMed Abstract:
The central region of MDM2 is critical for p53 activation and tumor suppression. Upon ribosomal stress, this region is bound by ribosomal proteins, particularly ribosomal protein L11 (RPL11), leading to MDM2 inactivation and subsequent p53 activation. Here, we solved the complex structure of human MDM2-RPL11 at 2.4 Å. MDM2 extensively interacts with RPL11 through an acidic domain and two zinc fingers. Formation of the MDM2-RPL11 complex induces substantial conformational changes in both proteins. RPL11, unable to bind MDM2 mutants, fails to induce the activation of p53 in cells. MDM2 mimics 28S rRNA binding to RPL11. The C4 zinc finger determines RPL11 binding to MDM2 but not its homolog, MDMX. Our results highlight the essential role of the RPL11-MDM2 interaction in p53 activation and tumor suppression and provide a structural basis for potential new anti-tumor drug development.
Organizational Affiliation:
State Key Laboratory of Agrobiotechnology, China Agricultural University, Beijing 100193, China;