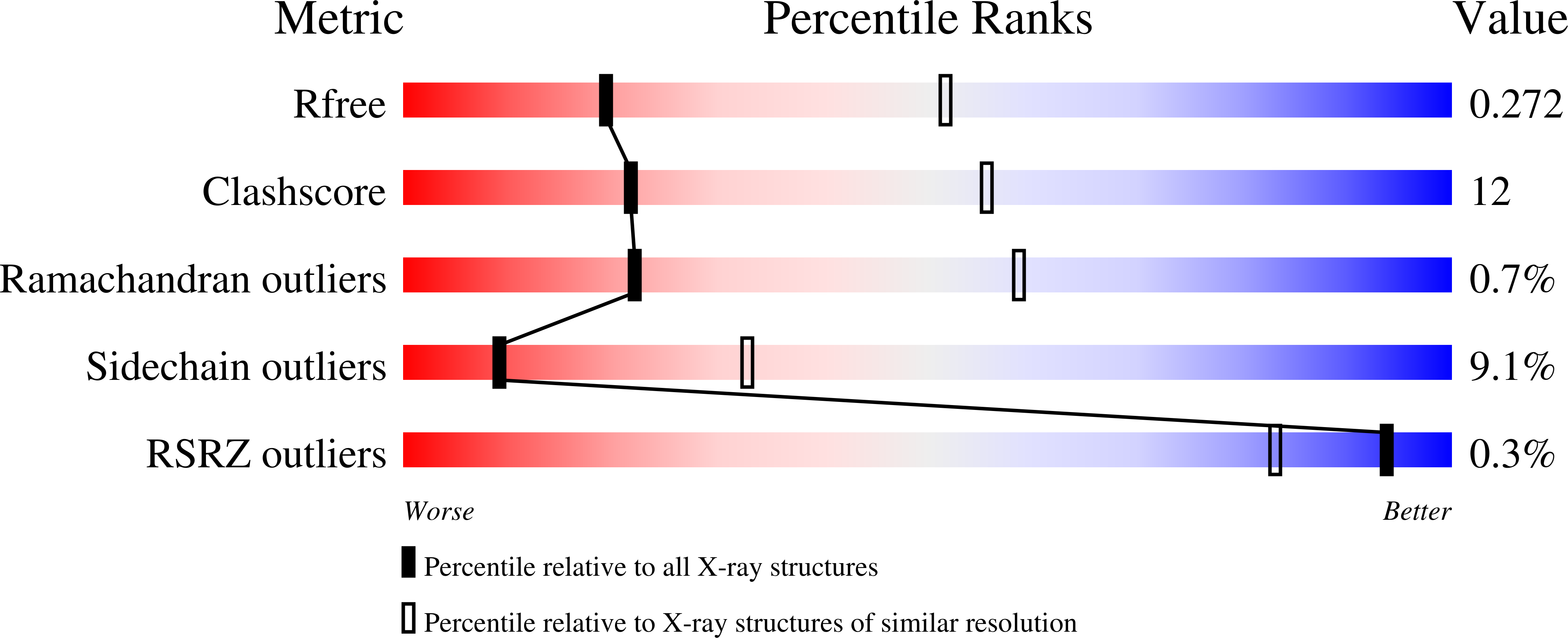
Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
Chamberlain, P.P., Wang, M., Riley, M., Delker, S., Carmel, G., Miller, K., Lopez-Girona, A., Pagarigan, B., Leon, B., Rychak, E., Corral, L., Lopez-Girona, A., Ren, Y., Wang, M., Riley, M., Delker, S., Ito, T., Hideki, A., Mori, T., Handa, H., Hakoshima, T., Daniel, T.O., Miller, K., Cathers, B.E., Carmel, G., Pagarigan, B., Leon, B., Rychak, E., Corral, L., Ren, Y.To be published.