Engineering of an isolated p110 alpha subunit of PI3K alpha permits crystallization and provides a platform for structure-based drug design.
Chen, P., Deng, Y.L., Bergqvist, S., Falk, M.D., Liu, W., Timofeevski, S., Brooun, A.(2014) Protein Sci 23: 1332-1340
- PubMed: 25043846
- DOI: https://doi.org/10.1002/pro.2517
- Primary Citation of Related Structures:
4TUU, 4TV3 - PubMed Abstract:
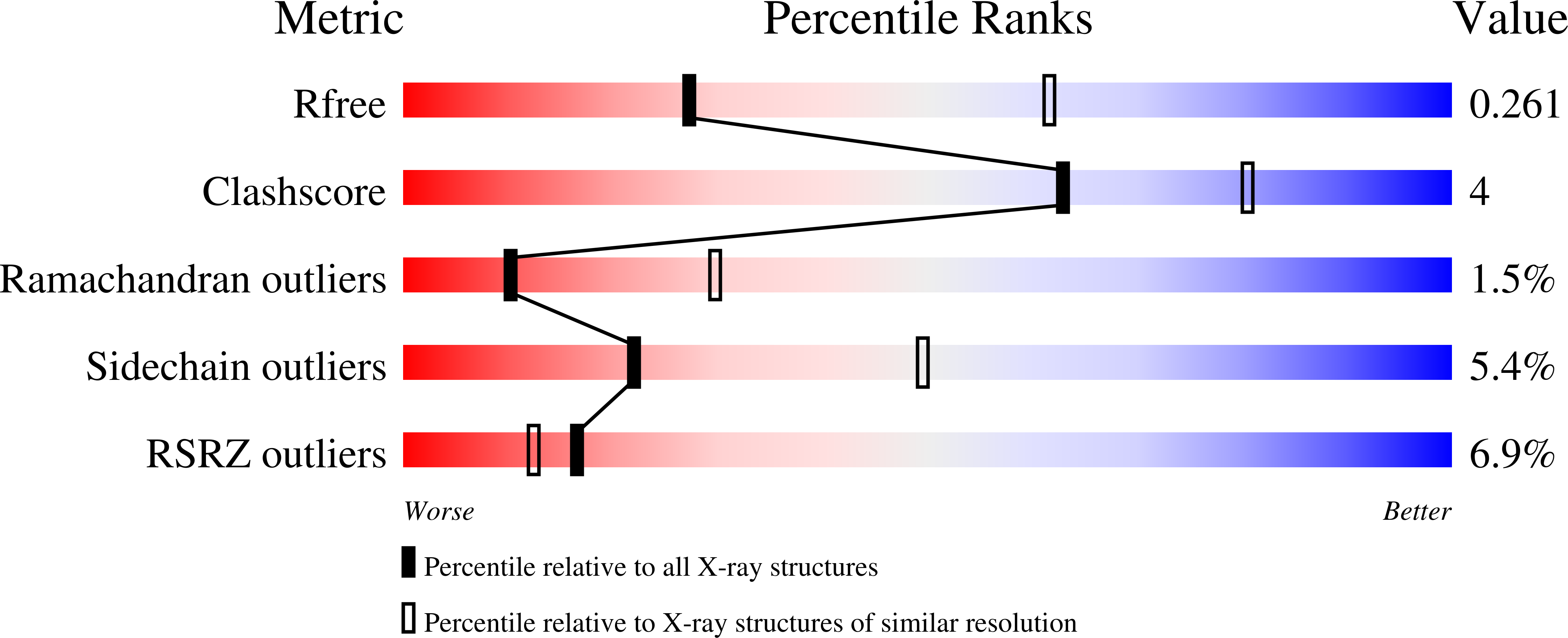
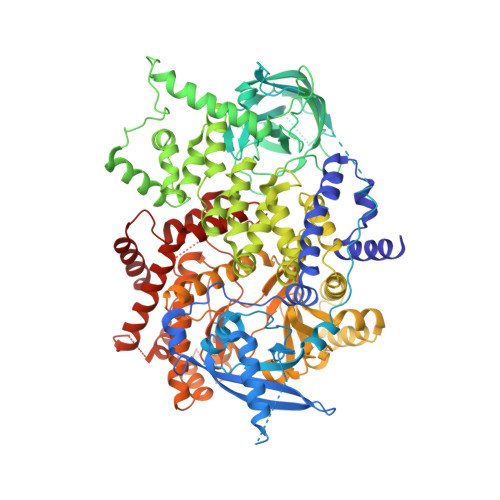
PI3Kα remains an attractive target for the development of anticancer targeted therapy. A number of p110α crystal structures in complex with the nSH2-iSH2 fragment of p85 regulatory subunit have been reported, including a few small molecule co-crystal structures, but the utilization of this crystal form is limited by low diffraction resolution and a crystal packing artifact that partially blocks the ATP binding site. Taking advantage of recent data on the functional characterization of the lipid binding properties of p110α, we designed a set of novel constructs allowing production of isolated stable p110α subunit missing the Adapter Binding Domain and lacking or featuring a modified C-terminal lipid binding motif. While this protein is not catalytically competent to phosphorylate its substrate PIP2, it retains ligand binding properties as indicated by direct binding studies with a pan-PI3Kα inhibitor. Additionally, we determined apo and PF-04691502 bound crystal structures of the p110α (105-1048) subunit at 2.65 and 2.85 Å, respectively. Comparison of isolated p110α(105-1048) with the p110α/p85 complex reveals a high degree of structural similarity, which validates suitability of this catalytically inactive p110α for iterative SBDD. Importantly, this crystal form of p110α readily accommodates the binding of noncovalent inhibitor by means of a fully accessible ATP site. The strategy presented here can be also applied to structural studies of other members of PI3KIA family.
Organizational Affiliation:
Oncology Structural Biology, Worldwide Research and Development, Pfizer Inc., San Diego, California, 92121.