Structural Basis for Different Substrate Profiles of Two Closely Related Class D beta-Lactamases and Their Inhibition by Halogens.
Stojanoski, V., Chow, D.C., Fryszczyn, B., Hu, L., Nordmann, P., Poirel, L., Sankaran, B., Prasad, B.V., Palzkill, T.(2015) Biochemistry 54: 3370-3380
- PubMed: 25938261
- DOI: https://doi.org/10.1021/acs.biochem.5b00298
- Primary Citation of Related Structures:
4S2L, 4S2M - PubMed Abstract:
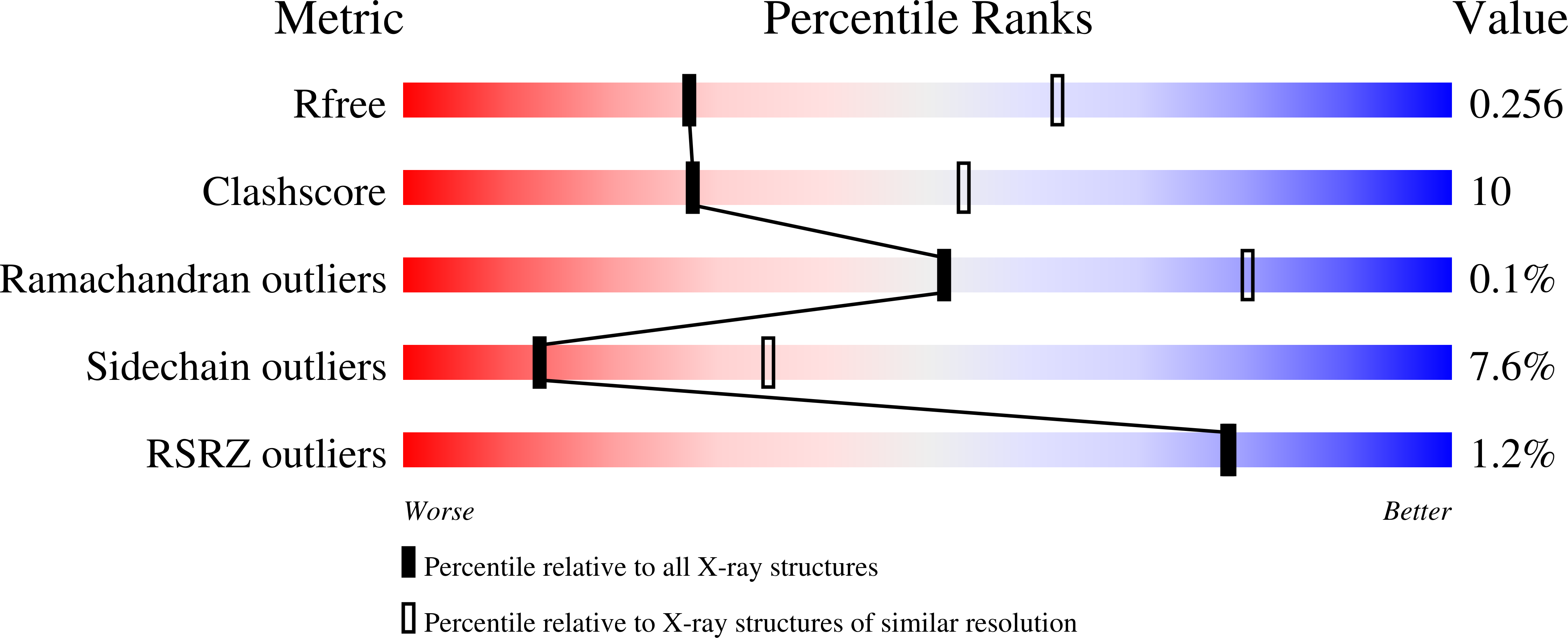
OXA-163 and OXA-48 are closely related class D β-lactamases that exhibit different substrate profiles. OXA-163 hydrolyzes oxyimino-cephalosporins, particularly ceftazidime, while OXA-48 prefers carbapenem substrates. OXA-163 differs from OXA-48 by one substitution (S212D) in the active-site β5 strand and a four-amino acid deletion (214-RIEP-217) in the loop connecting the β5 and β6 strands. Although the structure of OXA-48 has been determined, the structure of OXA-163 is unknown. To further understand the basis for their different substrate specificities, we performed enzyme kinetic analysis, inhibition assays, X-ray crystallography, and molecular modeling. The results confirm the carbapenemase nature of OXA-48 and the ability of OXA-163 to hydrolyze the oxyimino-cephalosporin ceftazidime. The crystal structure of OXA-163 determined at 1.72 Å resolution reveals an expanded active site compared to that of OXA-48, which allows the bulky substrate ceftazidime to be accommodated. The structural differences with OXA-48, which cannot hydrolyze ceftazidime, provide a rationale for the change in substrate specificity between the enzymes. OXA-163 also crystallized under another condition that included iodide. The crystal structure determined at 2.87 Å resolution revealed iodide in the active site accompanied by several significant conformational changes, including a distortion of the β5 strand, decarboxylation of Lys73, and distortion of the substrate-binding site. Further studies showed that both OXA-163 and OXA-48 are inhibited in the presence of iodide. In addition, OXA-10, which is not a member of the OXA-48-like family, is also inhibited by iodide. These findings provide a molecular basis for the hydrolysis of ceftazidime by OXA-163 and, more broadly, show how minor sequence changes can profoundly alter the active-site configuration and thereby affect the substrate profile of an enzyme.
Organizational Affiliation:
§Medical and Molecular Microbiology "Emerging Antibiotic Resistance" Unit, Department of Medicine, Faculty of Science, University of Fribourg, 1700 Fribourg, Switzerland.