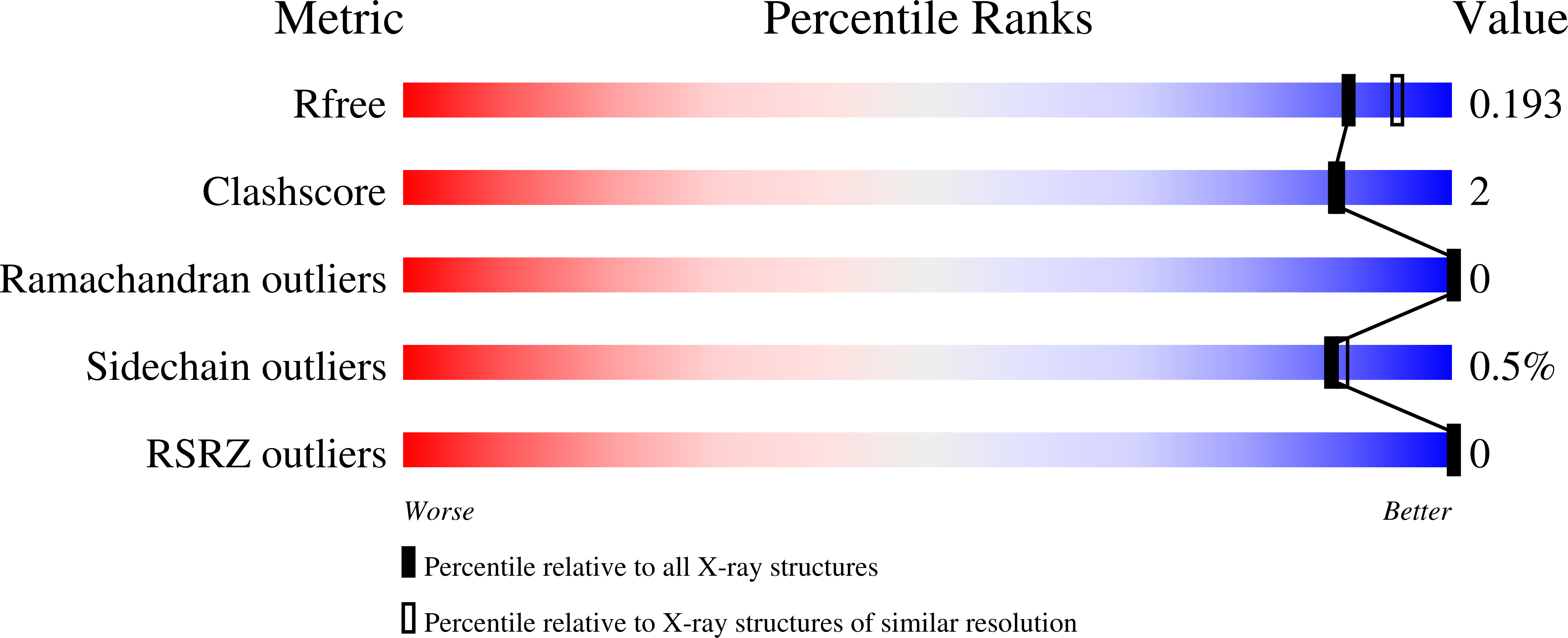
Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Ngo, A., Fong, K.T., Cox, D.L., Chen, X., Fisher, A.J.(2015) Acta Crystallogr D Biol Crystallogr 71: 1068-1076
- PubMed: 25945572
- DOI: https://doi.org/10.1107/S1399004715003326
- Primary Citation of Related Structures:
4R36, 4R37 - PubMed Abstract:
Uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (LpxA) catalyzes a reversible reaction for adding an O-acyl group to the GlcNAc in UDP-GlcNAc in the first step of lipid A biosynthesis. Lipid A constitutes a major component of lipopolysaccharides, also referred to as endotoxins, which form the outer monolayer of the outer membrane of Gram-negative bacteria. Ligand-free and UDP-GlcNAc-bound crystal structures of LpxA from Bacteroides fragilis NCTC 9343, the most common pathogenic bacteria found in abdominal abscesses, have been determined and are presented here. The enzyme crystallizes in a cubic space group, with the crystallographic threefold axis generating the biological functional homotrimer and with each monomer forming a nine-rung left-handed β-helical (LβH) fold in the N-terminus followed by an α-helical motif in the C-terminus. The structure is highly similar to LpxA from other organisms. Yet, despite sharing a similar LβH structure with LpxAs from Escherichia coli and others, previously unseen calcium ions are observed on the threefold axis in B. fragilis LpxA to help stabilize the trimeric assembly.
Organizational Affiliation:
Department of Chemistry, University of California, One Shields Avenue, Davis, CA 95616, USA.