Structural basis for the recruitment of glycogen synthase by glycogenin.
Zeqiraj, E., Tang, X., Hunter, R.W., Garcia-Rocha, M., Judd, A., Deak, M., von Wilamowitz-Moellendorff, A., Kurinov, I., Guinovart, J.J., Tyers, M., Sakamoto, K., Sicheri, F.(2014) Proc Natl Acad Sci U S A 111: E2831-E2840
- PubMed: 24982189
- DOI: https://doi.org/10.1073/pnas.1402926111
- Primary Citation of Related Structures:
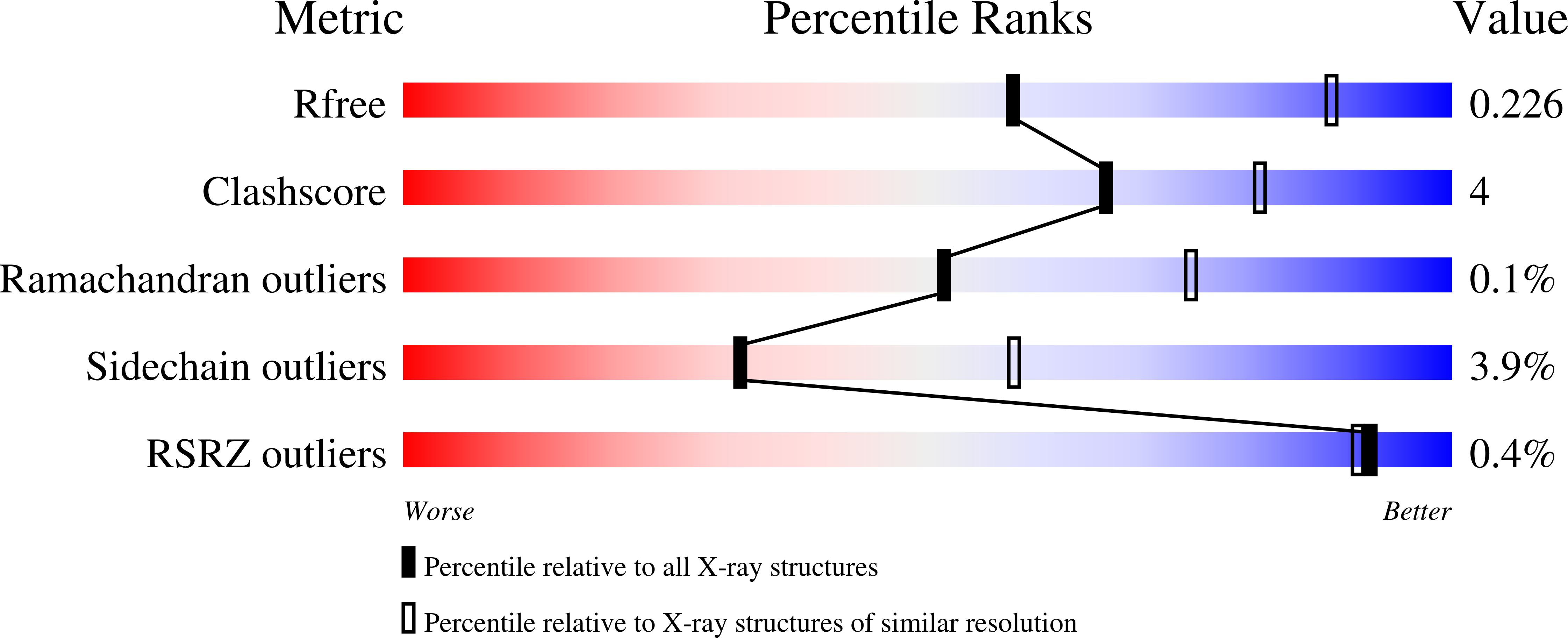
4QLB - PubMed Abstract:
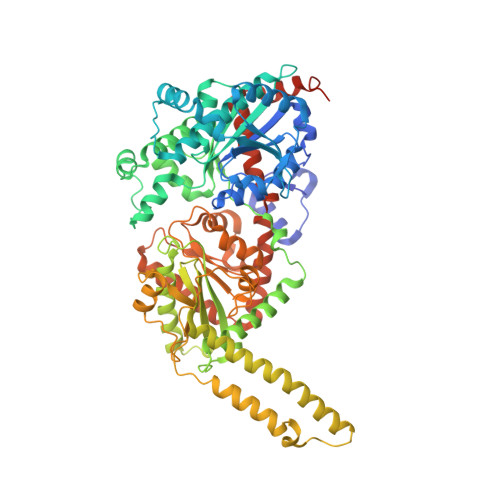
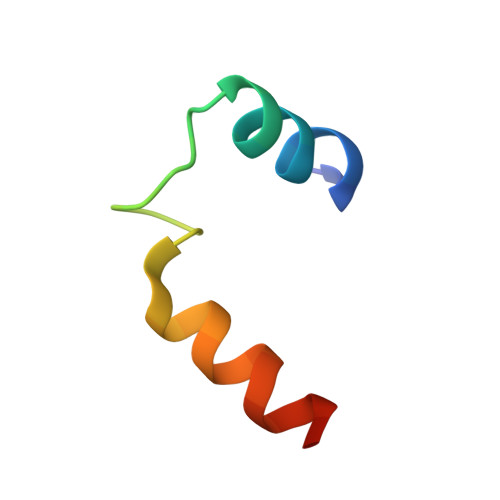
Glycogen is a primary form of energy storage in eukaryotes that is essential for glucose homeostasis. The glycogen polymer is synthesized from glucose through the cooperative action of glycogen synthase (GS), glycogenin (GN), and glycogen branching enzyme and forms particles that range in size from 10 to 290 nm. GS is regulated by allosteric activation upon glucose-6-phosphate binding and inactivation by phosphorylation on its N- and C-terminal regulatory tails. GS alone is incapable of starting synthesis of a glycogen particle de novo, but instead it extends preexisting chains initiated by glycogenin. The molecular determinants by which GS recognizes self-glucosylated GN, the first step in glycogenesis, are unknown. We describe the crystal structure of Caenorhabditis elegans GS in complex with a minimal GS targeting sequence in GN and show that a 34-residue region of GN binds to a conserved surface on GS that is distinct from previously characterized allosteric and binding surfaces on the enzyme. The interaction identified in the GS-GN costructure is required for GS-GN interaction and for glycogen synthesis in a cell-free system and in intact cells. The interaction of full-length GS-GN proteins is enhanced by an avidity effect imparted by a dimeric state of GN and a tetrameric state of GS. Finally, the structure of the N- and C-terminal regulatory tails of GS provide a basis for understanding phosphoregulation of glycogen synthesis. These results uncover a central molecular mechanism that governs glycogen metabolism.
Organizational Affiliation:
Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, ON, Canada M5G 1X5;