TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer.
Fayngerts, S.A., Wu, J., Oxley, C.L., Liu, X., Vourekas, A., Cathopoulis, T., Wang, Z., Cui, J., Liu, S., Sun, H., Lemmon, M.A., Zhang, L., Shi, Y., Chen, Y.H.(2014) Cancer Cell 26: 465-478
- PubMed: 25242044
- DOI: https://doi.org/10.1016/j.ccr.2014.07.025
- Primary Citation of Related Structures:
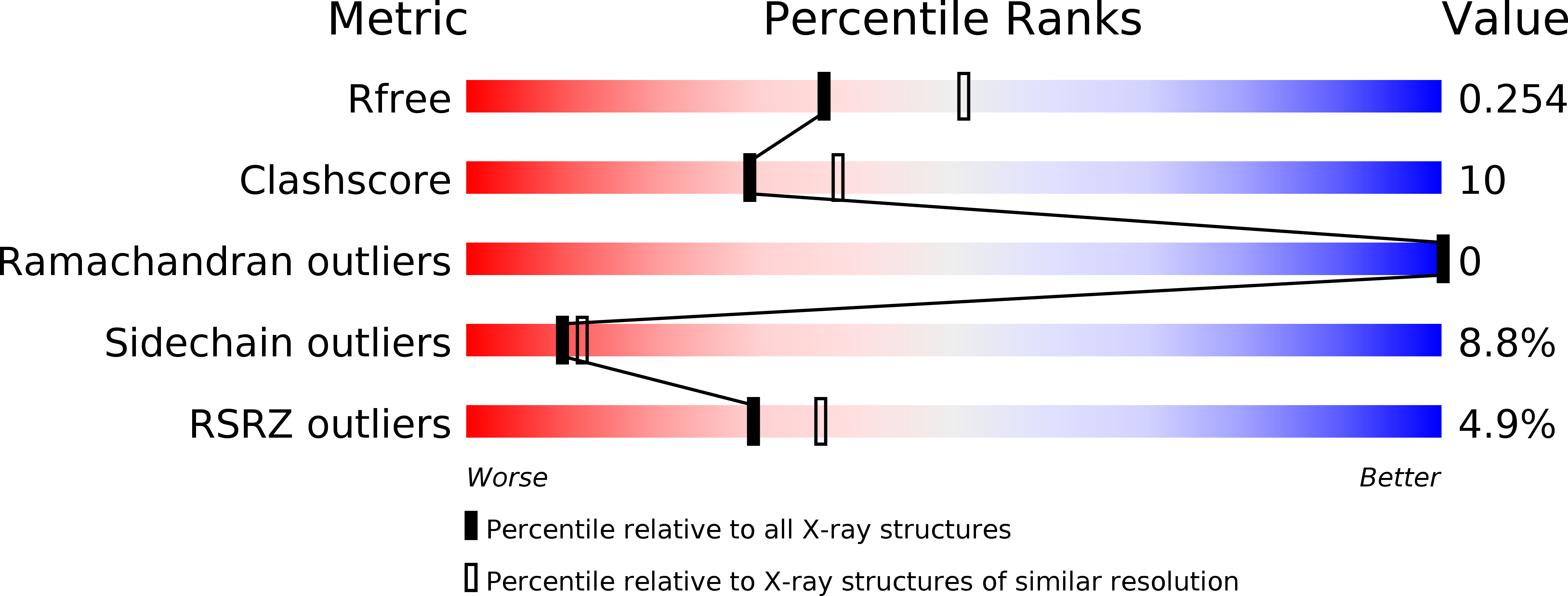
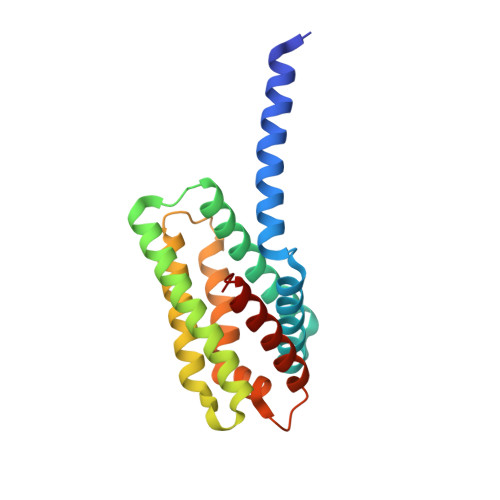
4Q9V - PubMed Abstract:
More than half of human cancers have aberrantly upregulated phosphoinositide signals; yet how phospholipid signals are controlled during tumorigenesis is not fully understood. We report here that TIPE3 (TNFAIP8L3) is the transfer protein of phosphoinositide second messengers that promote cancer. High-resolution crystal structure of TIPE3 shows a large hydrophobic cavity that is occupied by a phospholipid-like molecule. TIPE3 preferentially captures and shuttles two lipid second messengers, i.e., phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate, and increases their levels in the plasma membrane. Notably, human cancers have markedly upregulated TIPE3 expression. Knocking out TIPE3 diminishes tumorigenesis, whereas enforced TIPE3 expression enhances it in vivo. Thus, the function and metabolism of phosphoinositide second messengers are controlled by a specific transfer protein during tumorigenesis.
Organizational Affiliation:
Department of Pathology and Laboratory Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.