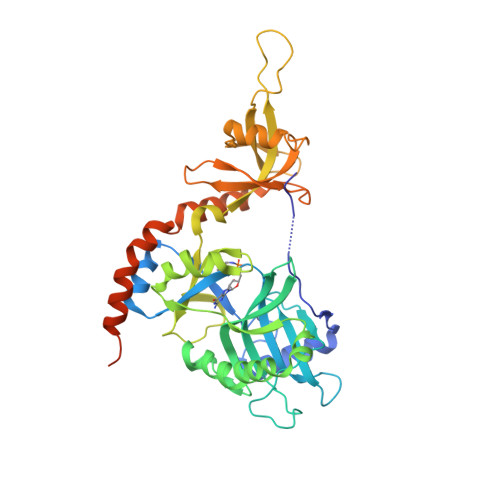
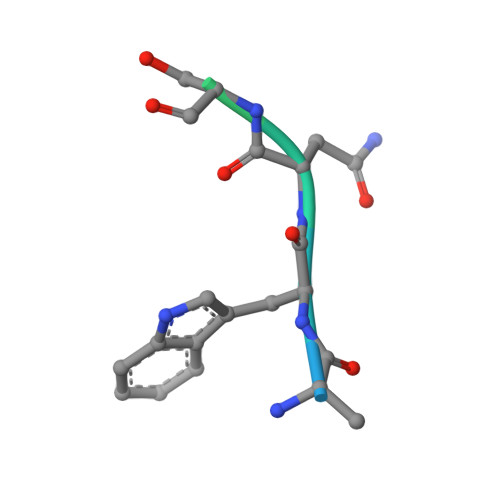
How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes.
Doamekpor, S.K., Sanchez, A.M., Schwer, B., Shuman, S., Lima, C.D.(2014) Genes Dev 28: 1323-1336
- PubMed: 24939935
- DOI: https://doi.org/10.1101/gad.242768.114
- Primary Citation of Related Structures:
4PZ6, 4PZ7, 4PZ8 - PubMed Abstract:
Interactions between RNA guanylyltransferase (GTase) and the C-terminal domain (CTD) repeats of RNA polymerase II (Pol2) and elongation factor Spt5 are thought to orchestrate cotranscriptional capping of nascent mRNAs. The crystal structure of a fission yeast GTase•Pol2 CTD complex reveals a unique docking site on the nucleotidyl transferase domain for an 8-amino-acid Pol2 CTD segment, S5PPSYSPTS5P, bracketed by two Ser5-PO4 marks. Analysis of GTase mutations that disrupt the Pol2 CTD interface shows that at least one of the two Ser5-PO4-binding sites is required for cell viability and that each site is important for cell growth at 37°C. Fission yeast GTase binds the Spt5 CTD at a separate docking site in the OB-fold domain that captures the Trp4 residue of the Spt5 nonapeptide repeat T(1)PAW(4)NSGSK. A disruptive mutation in the Spt5 CTD-binding site of GTase is synthetically lethal with mutations in the Pol2 CTD-binding site, signifying that the Spt5 and Pol2 CTDs cooperate to recruit capping enzyme in vivo. CTD phosphorylation has opposite effects on the interaction of GTase with Pol2 (Ser5-PO4 is required for binding) versus Spt5 (Thr1-PO4 inhibits binding). We propose that the state of Thr1 phosphorylation comprises a binary "Spt5 CTD code" that is read by capping enzyme independent of and parallel to its response to the state of the Pol2 CTD.
Organizational Affiliation:
Structural Biology Program, Sloan-Kettering Institute, New York, New York 10065, USA;