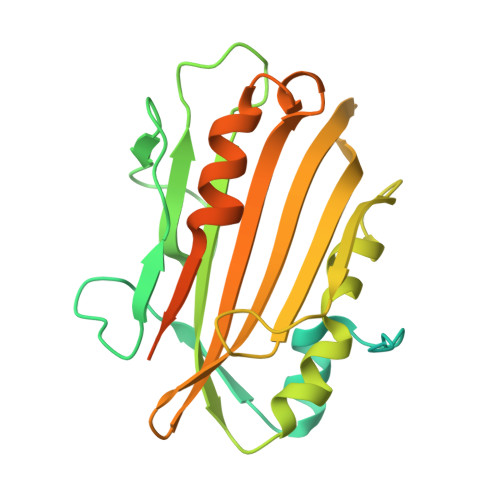
Structure-based Epitope Mapping of Mycobacterium tuberculosis Secretary Antigen MTC28
Kundu, P., Biswas, R., Mukherjee, S., Reinhard, L., Dutta, A., Mueller-Dieckmann, J., Weiss, M.S., Pal, N.K., Das, A.K.(2016) J Biol Chem 291: 13943-13954
- PubMed: 27189947
- DOI: https://doi.org/10.1074/jbc.M116.726422
- Primary Citation of Related Structures:
4OL4, 4PWS - PubMed Abstract:
Secretary proteins of Mycobacterium tuberculosis are key players of the mycobacterial infection pathway. MTC28 is a 28-kDa proline-rich secretary antigen of Mycobacterium tuberculosis and is only conserved in pathogenic strains of mycobacteria. Here we report the crystal structure of MTC28 at 2.8- and 2.15-Å resolutions for the structure-based epitope design. MTC28 shares a "mog1p"-fold consisting of seven antiparallel β strands stacked between α helices. Five probable epitopes have been located on a solvent-accessible flexible region by computational analysis of the structure of MTC28. Simultaneously, the protein is digested with trypsin and the resulting fragments are purified by HPLC. Such 10 purified peptide fragments are screened against sera from patients infected with pulmonary tuberculosis (PTB). Two of these 10 fragments, namely (127)ALDITLPMPPR(137) and (138)WTQVPDPNVPDAFVVIADR(156),are found to be major immunogenic epitopes that are localized on the outer surface of the protein molecule and are part of a single continuous epitope that have been predicted in silico Mutagenesis and antibody inhibition studies are in accordance with the results obtained from epitope mapping.
Organizational Affiliation:
Department of Biotechnology, Indian Institute of Technology Kharagpur, West Bengal 721302, India.