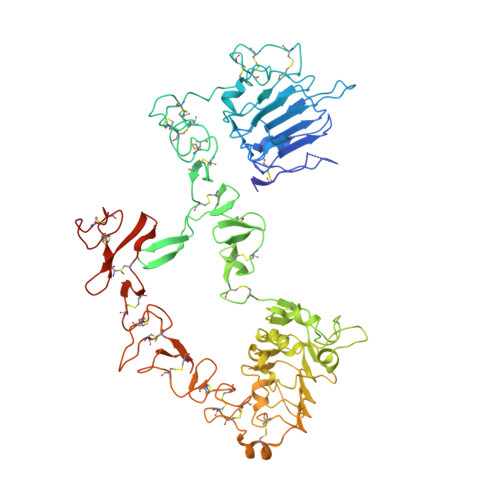
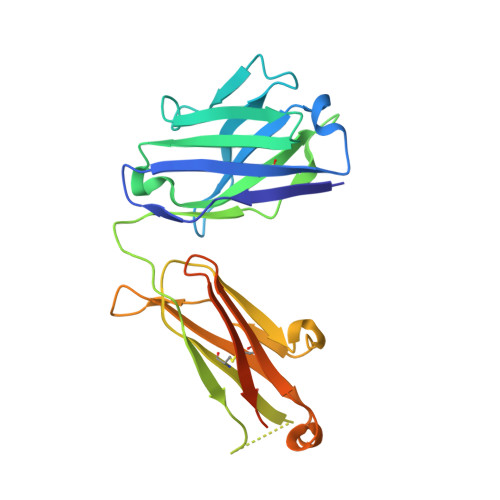
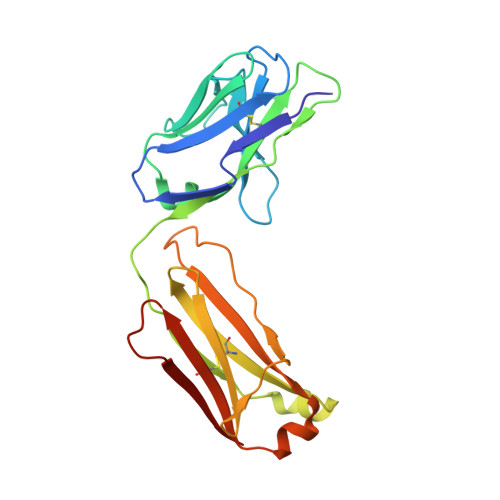
An antibody that locks HER3 in the inactive conformation inhibits tumor growth driven by HER2 or neuregulin.
Garner, A.P., Bialucha, C.U., Sprague, E.R., Garrett, J.T., Sheng, Q., Li, S., Sineshchekova, O., Saxena, P., Sutton, C.R., Chen, D., Chen, Y., Wang, H., Liang, J., Das, R., Mosher, R., Gu, J., Huang, A., Haubst, N., Zehetmeier, C., Haberl, M., Elis, W., Kunz, C., Heidt, A.B., Herlihy, K., Murtie, J., Schuller, A., Arteaga, C.L., Sellers, W.R., Ettenberg, S.A.(2013) Cancer Res 73: 6024-6035
- PubMed: 23928993
- DOI: https://doi.org/10.1158/0008-5472.CAN-13-1198
- Primary Citation of Related Structures:
4P59 - PubMed Abstract:
HER2/HER3 dimerization resulting from overexpression of HER2 or neuregulin (NRG1) in cancer leads to HER3-mediated oncogenic activation of phosphoinositide 3-kinase (PI3K) signaling. Although ligand-blocking HER3 antibodies inhibit NRG1-driven tumor growth, they are ineffective against HER2-driven tumor growth because HER2 activates HER3 in a ligand-independent manner. In this study, we describe a novel HER3 monoclonal antibody (LJM716) that can neutralize multiple modes of HER3 activation, making it a superior candidate for clinical translation as a therapeutic candidate. LJM716 was a potent inhibitor of HER3/AKT phosphorylation and proliferation in HER2-amplified and NRG1-expressing cancer cells, and it displayed single-agent efficacy in tumor xenograft models. Combining LJM716 with agents that target HER2 or EGFR produced synergistic antitumor activity in vitro and in vivo. In particular, combining LJM716 with trastuzumab produced a more potent inhibition of signaling and cell proliferation than trastuzumab/pertuzumab combinations with similar activity in vivo. To elucidate its mechanism of action, we solved the structure of LJM716 bound to HER3, finding that LJM716 bound to an epitope, within domains 2 and 4, that traps HER3 in an inactive conformation. Taken together, our findings establish that LJM716 possesses a novel mechanism of action that, in combination with HER2- or EGFR-targeted agents, may leverage their clinical efficacy in ErbB-driven cancers.
Organizational Affiliation:
Authors' Affiliations: Novartis Institutes for Biomedical Research, Cambridge, Massachusetts; Departments of Medicine and Cancer Biology; Breast Cancer Research Program, Vanderbilt-Ingram Cancer Center, Vanderbilt University, Nashville, Tennessee; MorphoSys AG, Martinsried, Germany; and Genomics Institute of the Novartis Research Foundation, San Diego, California.