Broad-spectrum allosteric inhibition of herpesvirus proteases.
Gable, J.E., Lee, G.M., Jaishankar, P., Hearn, B.R., Waddling, C.A., Renslo, A.R., Craik, C.S.(2014) Biochemistry 53: 4648-4660
- PubMed: 24977643
- DOI: https://doi.org/10.1021/bi5003234
- Primary Citation of Related Structures:
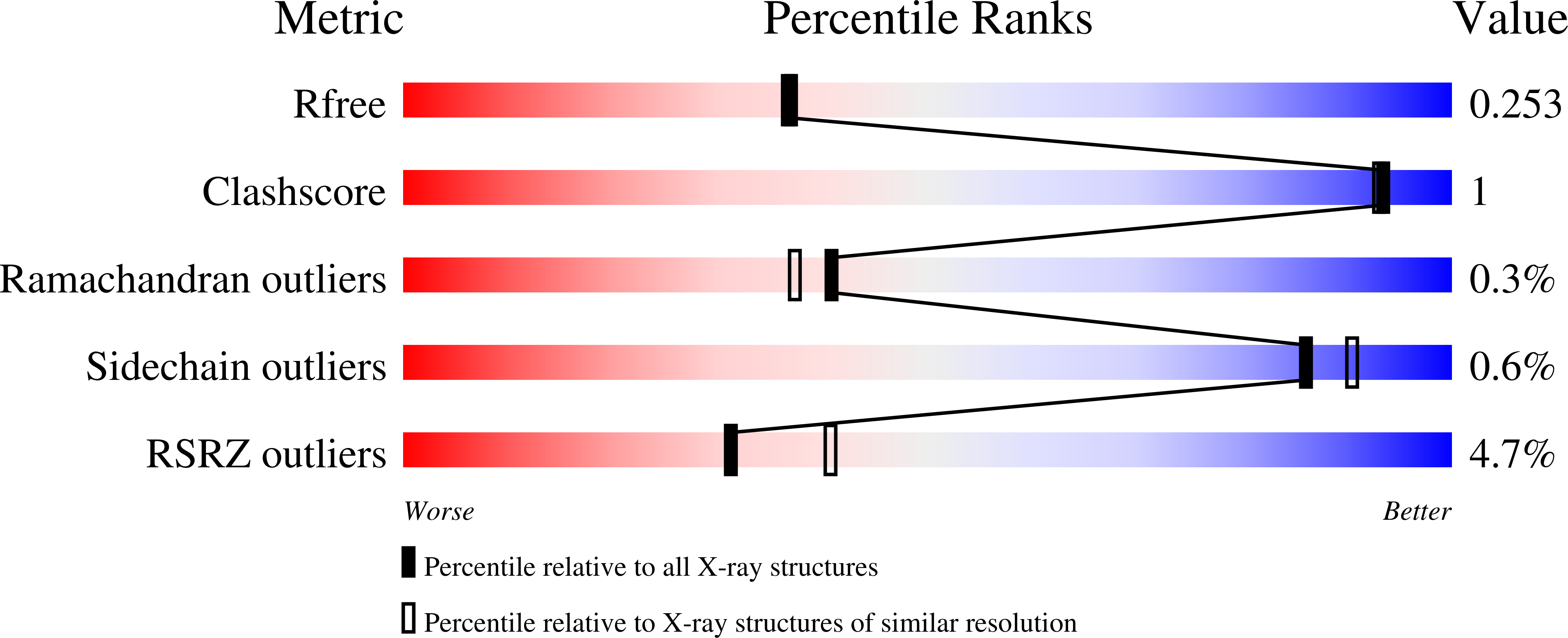
4P2T, 4P3H - PubMed Abstract:
Herpesviruses rely on a homodimeric protease for viral capsid maturation. A small molecule, DD2, previously shown to disrupt dimerization of Kaposi's sarcoma-associated herpesvirus protease (KSHV Pr) by trapping an inactive monomeric conformation and two analogues generated through carboxylate bioisosteric replacement (compounds 2 and 3) were shown to inhibit the associated proteases of all three human herpesvirus (HHV) subfamilies (α, β, and γ). Inhibition data reveal that compound 2 has potency comparable to or better than that of DD2 against the tested proteases. Nuclear magnetic resonance spectroscopy and a new application of the kinetic analysis developed by Zhang and Poorman [Zhang, Z. Y., Poorman, R. A., et al. (1991) J. Biol. Chem. 266, 15591-15594] show DD2, compound 2, and compound 3 inhibit HHV proteases by dimer disruption. All three compounds bind the dimer interface of other HHV proteases in a manner analogous to binding of DD2 to KSHV protease. The determination and analysis of cocrystal structures of both analogues with the KSHV Pr monomer verify and elaborate on the mode of binding for this chemical scaffold, explaining a newly observed critical structure-activity relationship. These results reveal a prototypical chemical scaffold for broad-spectrum allosteric inhibition of human herpesvirus proteases and an approach for the identification of small molecules that allosterically regulate protein activity by targeting protein-protein interactions.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California , San Francisco, California 94158-2280, United States.