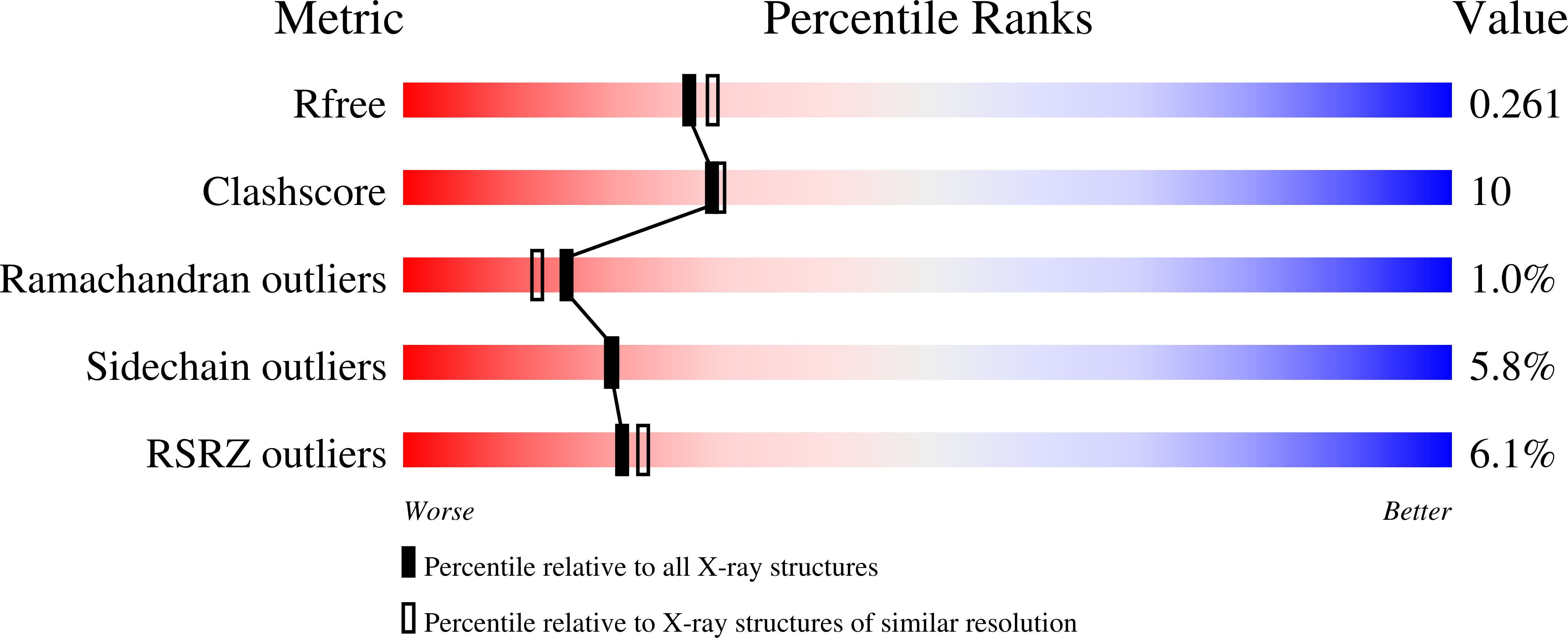
Allostery and Conformational Dynamics in cAMP-binding Acyltransferases.
Podobnik, M., Siddiqui, N., Rebolj, K., Nambi, S., Merzel, F., Visweswariah, S.S.(2014) J Biol Chem 289: 16588-16600
- PubMed: 24748621
- DOI: https://doi.org/10.1074/jbc.M114.560086
- Primary Citation of Related Structures:
4OLL, 4ONU, 4ORF - PubMed Abstract:
Mycobacteria harbor unique proteins that regulate protein lysine acylation in a cAMP-regulated manner. These lysine acyltransferases from Mycobacterium smegmatis (KATms) and Mycobacterium tuberculosis (KATmt) show distinctive biochemical properties in terms of cAMP binding affinity to the N-terminal cyclic nucleotide binding domain and allosteric activation of the C-terminal acyltransferase domain. Here we provide evidence for structural features in KATms that account for high affinity cAMP binding and elevated acyltransferase activity in the absence of cAMP. Structure-guided mutational analysis converted KATms from a cAMP-regulated to a cAMP-dependent acyltransferase and identified a unique asparagine residue in the acyltransferase domain of KATms that assists in the enzymatic reaction in the absence of a highly conserved glutamate residue seen in Gcn5-related N-acetyltransferase-like acyltransferases. Thus, we have identified mechanisms by which properties of similar proteins have diverged in two species of mycobacteria by modifications in amino acid sequence, which can dramatically alter the abundance of conformational states adopted by a protein.
Organizational Affiliation:
From the Laboratory for Molecular Biology and Nanobiotechnology and marjetka.podobnik@ki.si.