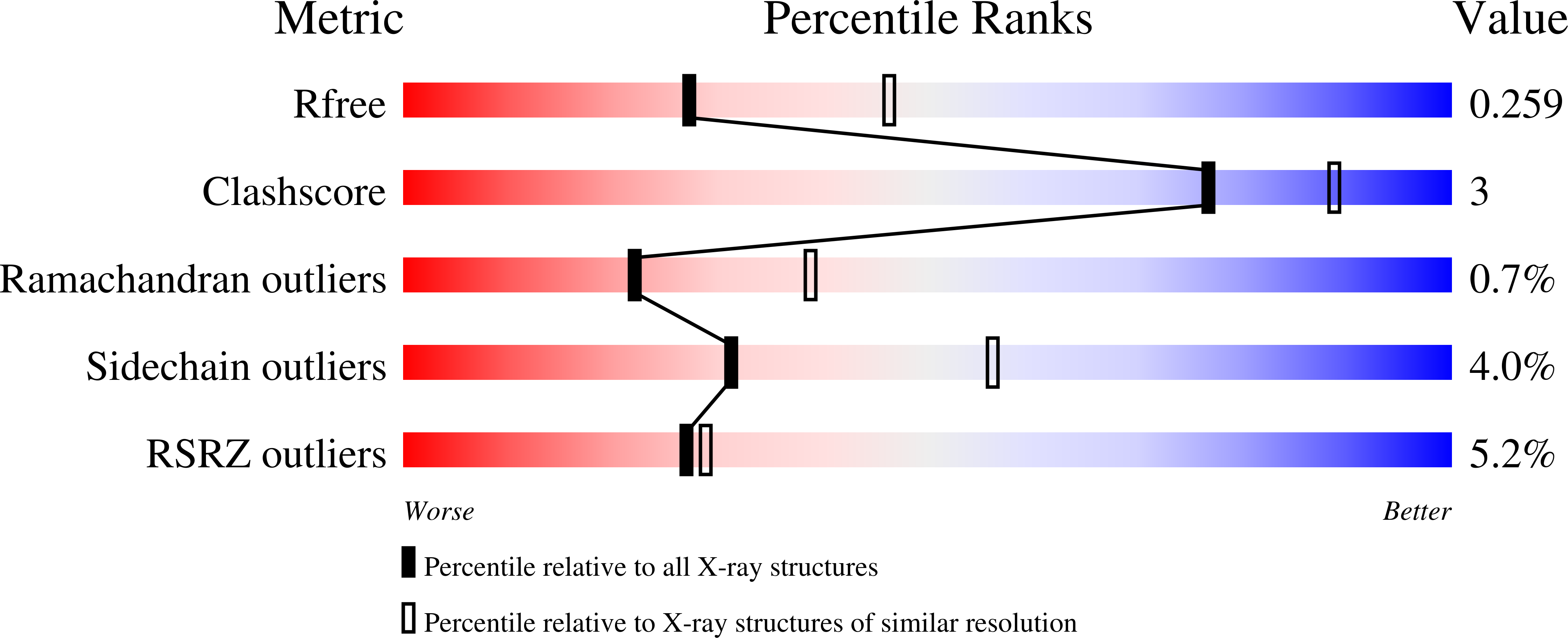
Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Persch, E., Bryson, S., Todoroff, N.K., Eberle, C., Thelemann, J., Dirdjaja, N., Kaiser, M., Weber, M., Derbani, H., Brun, R., Schneider, G., Pai, E.F., Krauth-Siegel, R.L., Diederich, F.(2014) ChemMedChem 9: 1880-1891
- PubMed: 24788386
- DOI: https://doi.org/10.1002/cmdc.201402032
- Primary Citation of Related Structures:
4NEV, 4NEW - PubMed Abstract:
The causative agents of the parasitic disease human African trypanosomiasis belong to the family of trypanosomatids. These parasitic protozoa exhibit a unique thiol redox metabolism that is based on the flavoenzyme trypanothione reductase (TR). TR was identified as a potential drug target and features a large active site that allows a multitude of possible ligand orientations, which renders rational structure-based inhibitor design highly challenging. Herein we describe the synthesis, binding properties, and kinetic analysis of a new series of small-molecule inhibitors of TR. The conjunction of biological activities, mutation studies, and virtual ligand docking simulations led to the prediction of a binding mode that was confirmed by crystal structure analysis. The crystal structures revealed that the ligands bind to the hydrophobic wall of the so-called "mepacrine binding site". The binding conformation and potency of the inhibitors varied for TR from Trypanosoma brucei and T. cruzi.
Organizational Affiliation:
Laboratorium für Organische Chemie, ETH Zürich, Vladimir-Prelog-Weg 3, 8093 Zurich (Switzerland).