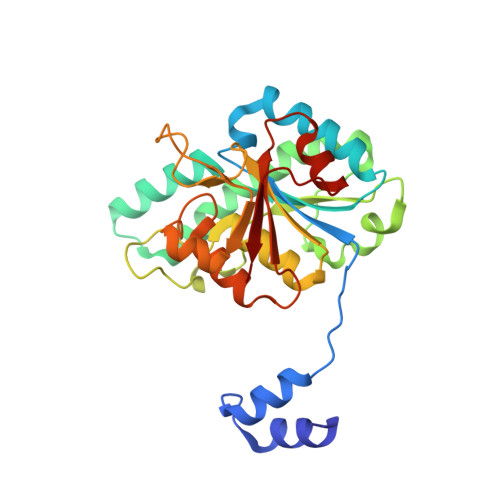
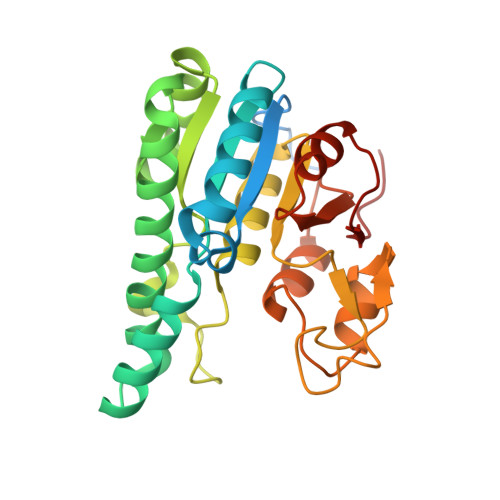
Structural diversity of polyoxomolybdate clusters along the three-fold axis of the molybdenum storage protein.
Poppe, J., Warkentin, E., Demmer, U., Kowalewski, B., Dierks, T., Schneider, K., Ermler, U.(2014) J Inorg Biochem 138: 122-128
- PubMed: 24945101
- DOI: https://doi.org/10.1016/j.jinorgbio.2014.05.009
- Primary Citation of Related Structures:
4NDO, 4NDP, 4NDQ, 4NDR - PubMed Abstract:
The molybdenum storage protein (MoSto) can store more than 100 Mo or W atoms as discrete polyoxometalate (POM) clusters. Here, we describe the three POM cluster sites along the threefold axis of the protein complex based on four X-ray structures with slightly different polyoxomolybdate compositions between 1.35 and 2 Å resolution. In contrast to the Moα-out binding site occupied by an Mo3 cluster, the Moα-in and Moβ binding sites contain rather weak and non-uniform electron density for the Mo atoms (but clearly identifiable by anomalous data), suggesting the presence of POM cluster ensembles and/or degradation products of larger aggregates. The "Moα-in cluster ensemble" was interpreted as an antiprism-like Mo6 species superimposed with an Mo7 pyramide and the "Moβ cluster ensemble" as an Mo13 cluster (present mostly in a degraded form) composed of a pyramidal Mo7 and a Mo3 building block linked by three spatially separated MoOx units. Inside the ball-shaped Mo13 cluster sits an occluded central atom, perhaps a metal ion. POM cluster formation at the Moα-in and Moβ sites appears to be driven by filtering out and binding/protecting self-assembled transient species complementary to the protein template.
Organizational Affiliation:
Max-Planck-Institut für Biophysik, Max-von-Laue-Straße 3, D-60438 Frankfurt am Main, Germany.