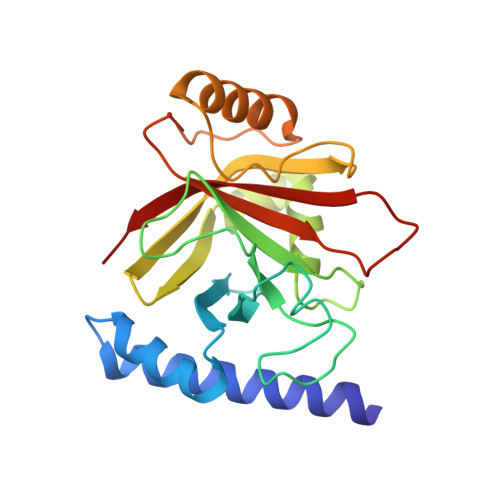
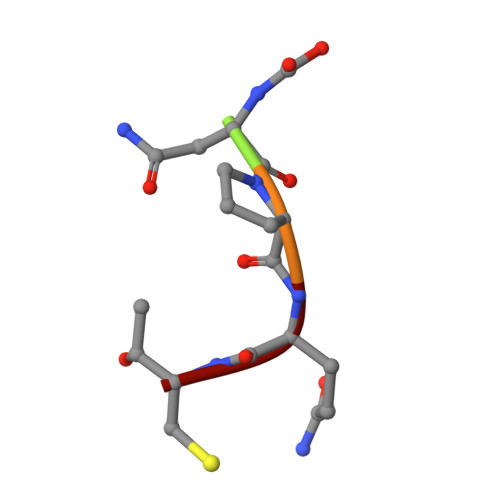
Structural and Computational Studies of the Staphylococcus aureus Sortase B-Substrate Complex Reveal a Substrate-stabilized Oxyanion Hole.
Jacobitz, A.W., Wereszczynski, J., Yi, S.W., Amer, B.R., Huang, G.L., Nguyen, A.V., Sawaya, M.R., Jung, M.E., McCammon, J.A., Clubb, R.T.(2014) J Biol Chem 289: 8891-8902
- PubMed: 24519933
- DOI: https://doi.org/10.1074/jbc.M113.509273
- Primary Citation of Related Structures:
4LFD - PubMed Abstract:
Sortase cysteine transpeptidases covalently attach proteins to the bacterial cell wall or assemble fiber-like pili that promote bacterial adhesion. Members of this enzyme superfamily are widely distributed in Gram-positive bacteria that frequently utilize multiple sortases to elaborate their peptidoglycan. Sortases catalyze transpeptidation using a conserved active site His-Cys-Arg triad that joins a sorting signal located at the C terminus of their protein substrate to an amino nucleophile located on the cell surface. However, despite extensive study, the catalytic mechanism and molecular basis of substrate recognition remains poorly understood. Here we report the crystal structure of the Staphylococcus aureus sortase B enzyme in a covalent complex with an analog of its NPQTN sorting signal substrate, revealing the structural basis through which it displays the IsdC protein involved in heme-iron scavenging from human hemoglobin. The results of computational modeling, molecular dynamics simulations, and targeted amino acid mutagenesis indicate that the backbone amide of Glu(224) and the side chain of Arg(233) form an oxyanion hole in sortase B that stabilizes high energy tetrahedral catalytic intermediates. Surprisingly, a highly conserved threonine residue within the bound sorting signal substrate facilitates construction of the oxyanion hole by stabilizing the position of the active site arginine residue via hydrogen bonding. Molecular dynamics simulations and primary sequence conservation suggest that the sorting signal-stabilized oxyanion hole is a universal feature of enzymes within the sortase superfamily.
Organizational Affiliation:
From the Department of Chemistry and Biochemistry.