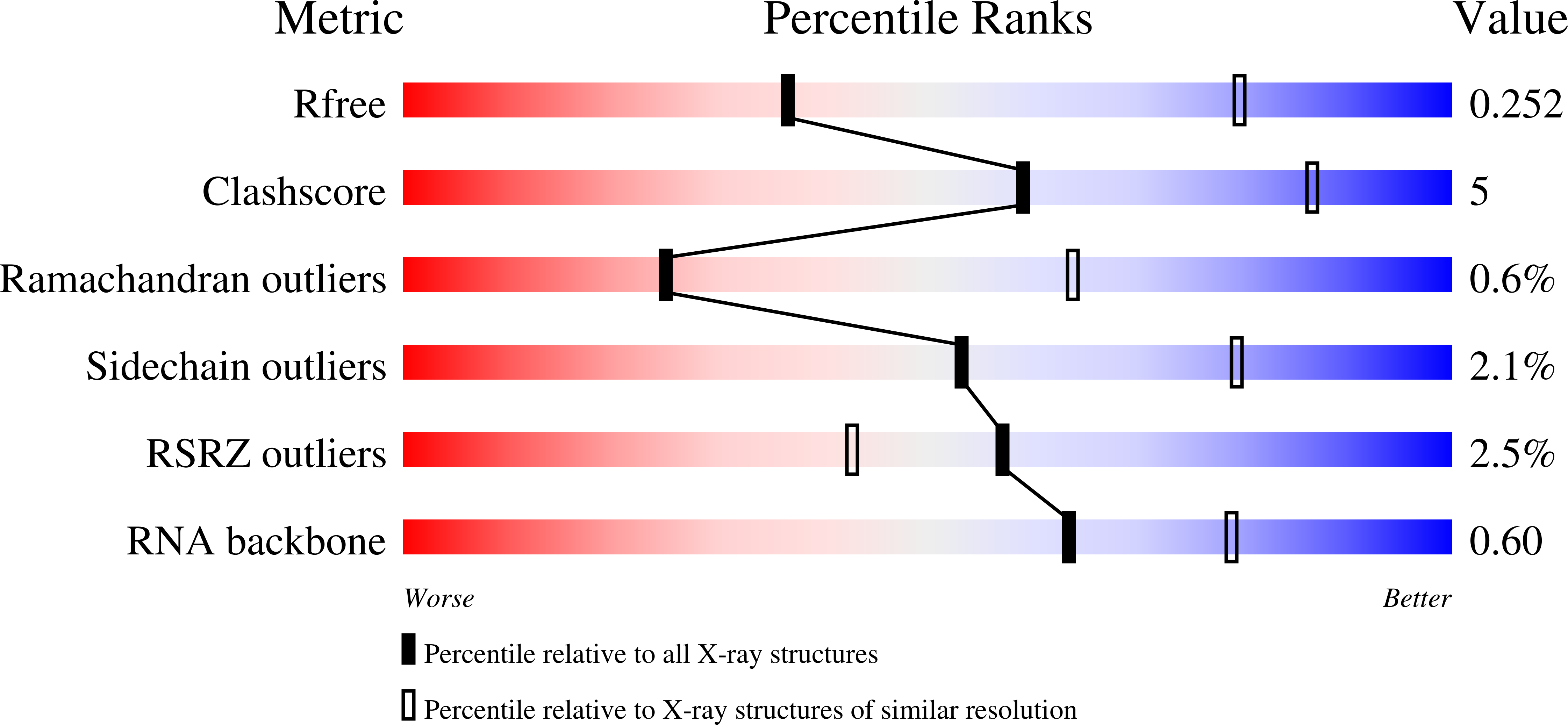
Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA.
Zhang, J., Ferre-D'Amare, A.R.(2013) Nature 500: 363-366
- PubMed: 23892783
- DOI: https://doi.org/10.1038/nature12440
- Primary Citation of Related Structures:
4LCK - PubMed Abstract:
In Gram-positive bacteria, T-box riboswitches regulate the expression of aminoacyl-tRNA synthetases and other proteins in response to fluctuating transfer RNA aminoacylation levels under various nutritional states. T-boxes reside in the 5'-untranslated regions of the messenger RNAs they regulate, and consist of two conserved domains. Stem I contains the specifier trinucleotide that base pairs with the anticodon of cognate tRNA. 3' to stem I is the antiterminator domain, which base pairs with the tRNA acceptor end and evaluates its aminoacylation state. Despite high phylogenetic conservation and widespread occurrence in pathogens, the structural basis of tRNA recognition by this riboswitch remains ill defined. Here we demonstrate that the ~100-nucleotide T-box stem I is necessary and sufficient for specific, high-affinity (dissociation constant (Kd) ~150 nM) tRNA binding, and report the structure of Oceanobacillus iheyensis glyQ stem I in complex with its cognate tRNA at 3.2 Å resolution. Stem I recognizes the overall architecture of tRNA in addition to its anticodon, something accomplished by large ribonucleoproteins such as the ribosome, or proteins such as aminoacyl-tRNA synthetases, but is unprecedented for a compact mRNA domain. The C-shaped stem I cradles the L-shaped tRNA, forming an extended (1,604 Å(2)) intermolecular interface. In addition to the specifier-anticodon interaction, two interdigitated T-loops near the apex of stem I stack on the tRNA elbow in a manner analogous to those of the J11/12-J12/11 motif of RNase P and the L1 stalk of the ribosomal E-site. Because these ribonucleoproteins and T-boxes are unrelated, this strategy to recognize a universal tRNA feature probably evolved convergently. Mutually induced fit of stem I and the tRNA exploiting the intrinsic flexibility of tRNA and its conserved post-transcriptional modifications results in high shape complementarity, which in addition to providing specificity and affinity, globally organizes the T-box to orchestrate tRNA-dependent transcription regulation.
Organizational Affiliation:
National Heart, Lung and Blood Institute, 50 South Drive, MSC 8012, Bethesda, Maryland 20892-8012, USA.