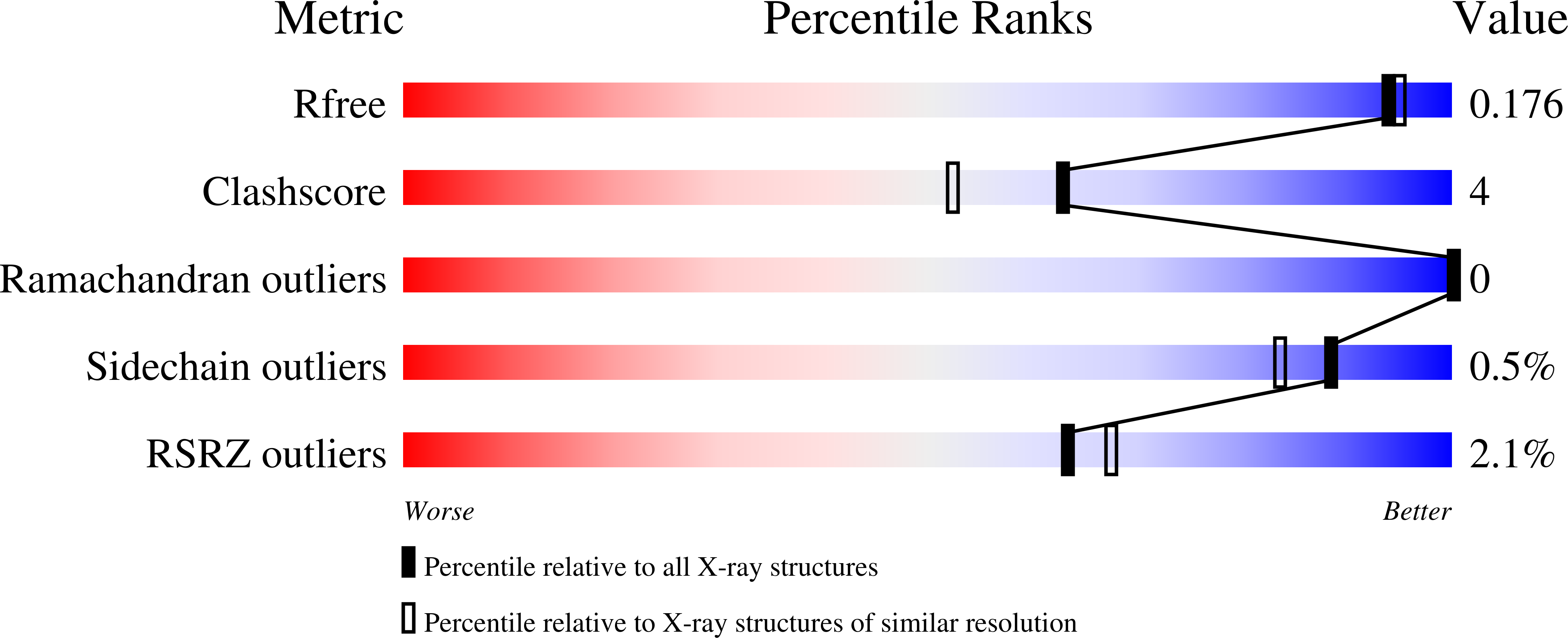
Structural and Functional Characterization of MppR, an Enduracididine Biosynthetic Enzyme from Streptomyces hygroscopicus: Functional Diversity in the Acetoacetate Decarboxylase-like Superfamily.
Burroughs, A.M., Hoppe, R.W., Goebel, N.C., Sayyed, B.H., Voegtline, T.J., Schwabacher, A.W., Zabriskie, T.M., Silvaggi, N.R.(2013) Biochemistry 52: 4492-4506
- PubMed: 23758195
- DOI: https://doi.org/10.1021/bi400397k
- Primary Citation of Related Structures:
4JM3, 4JMC, 4JMD, 4JME - PubMed Abstract:
The nonproteinogenic amino acid enduracididine is a critical component of the mannopeptimycins, cyclic glycopeptide antibiotics with activity against drug-resistant pathogens, including methicillin-resistant Staphylococcus aureus. Enduracididine is produced in Streptomyces hygroscopicus by three enzymes, MppP, MppQ, and MppR. On the basis of primary sequence analysis, MppP and MppQ are pyridoxal 5'-phosphate-dependent aminotransferases; MppR shares a low, but significant, level of sequence identity with acetoacetate decarboxylase. The exact reactions catalyzed by each enzyme and the intermediates involved in the route to enduracididine are currently unknown. Herein, we present biochemical and structural characterization of MppR that demonstrates a catalytic activity for this enzyme and provides clues about its role in enduracididine biosynthesis. Bioinformatic analysis shows that MppR belongs to a previously uncharacterized family within the acetoacetate decarboxylase-like superfamily (ADCSF) and suggests that MppR-like enzymes may catalyze reactions diverging from the well-characterized, prototypical ADCSF decarboxylase activity. MppR shares a high degree of structural similarity with acetoacetate decarboxylase, though the respective quaternary structures differ markedly and structural differences in the active site explain the observed loss of decarboxylase activity. The crystal structure of MppR in the presence of a mixture of pyruvate and 4-imidazolecarboxaldehyde shows that MppR catalyzes the aldol condensation of these compounds and subsequent dehydration. Surprisingly, the structure of MppR in the presence of "4-hydroxy-2-ketoarginine" shows the correct 4R enantiomer of "2-ketoenduracididine" bound to the enzyme. These data, together with bioinformatic analysis of MppR homologues, identify a novel family within the acetoacetate decarboxylase-like superfamily with divergent active site structure and, consequently, biochemical function.
Organizational Affiliation:
National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894, USA.