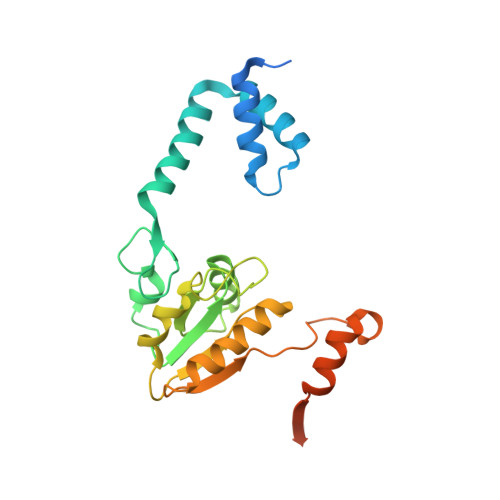
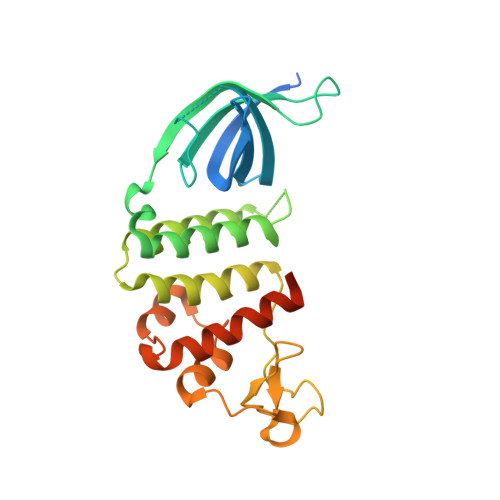
An 'open' structure of the RecOR complex supports ssDNA binding within the core of the complex.
Radzimanowski, J., Dehez, F., Round, A., Bidon-Chanal, A., McSweeney, S., Timmins, J.(2013) Nucleic Acids Res 41: 7972-7986
- PubMed: 23814185
- DOI: https://doi.org/10.1093/nar/gkt572
- Primary Citation of Related Structures:
4JCV - PubMed Abstract:
Efficient DNA repair is critical for cell survival and the maintenance of genome integrity. The homologous recombination pathway is responsible for the repair of DNA double-strand breaks within cells. Initiation of this pathway in bacteria can be carried out by either the RecBCD or the RecFOR proteins. An important regulatory player within the RecFOR pathway is the RecOR complex that facilitates RecA loading onto DNA. Here we report new data regarding the assembly of Deinococcus radiodurans RecOR and its interaction with DNA, providing novel mechanistic insight into the mode of action of RecOR in homologous recombination. We present a higher resolution crystal structure of RecOR in an 'open' conformation in which the tetrameric RecR ring flanked by two RecO molecules is accessible for DNA binding. We show using small-angle neutron scattering and mutagenesis studies that DNA binding does indeed occur within the RecR ring. Binding of single-stranded DNA occurs without any major conformational changes of the RecOR complex while structural rearrangements are observed on double-stranded DNA binding. Finally, our molecular dynamics simulations, supported by our biochemical data, provide a detailed picture of the DNA binding motif of RecOR and reveal that single-stranded DNA is sandwiched between the two facing oligonucleotide binding domains of RecO within the RecR ring.
Organizational Affiliation:
Structural Biology Group, European Synchrotron Radiation Facility, 6 rue Jules Horowitz, 38043 Grenoble cedex 9, France, Université de Lorraine, BP239, 54506 Vandoeuvre-lès-Nancy Cedex, France, CNRS, UMR N°7565, 54506 Vandoeuvre-les-Nancy, France, European Molecular Biology Laboratory, Grenoble Outstation, 6 rue Jules Horowitz, 38042 Grenoble, France, Unit for Virus Host-Cell Interactions, Univ. Grenoble Alpes-EMBL-CNRS, 6 rue Jules Horowitz, 38042 Grenoble, France and Institut de Biologie Structurale, CNRS/CEA/Université de Grenoble, 41 rue Jules Horowitz, 38027 Grenoble cedex 1, France.