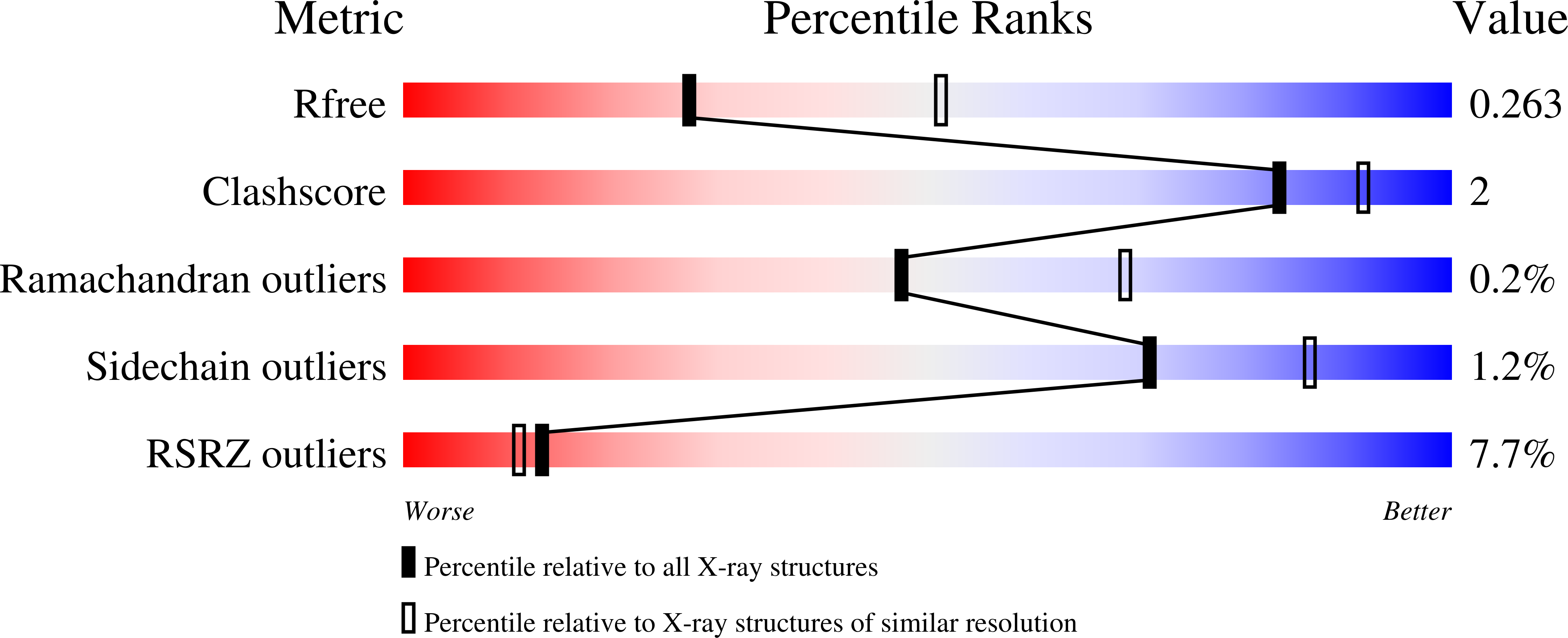
Metal binding properties of Escherichia coli YjiA, a member of the metal homeostasis-associated COG0523 family of GTPases.
Sydor, A.M., Jost, M., Ryan, K.S., Turo, K.E., Douglas, C.D., Drennan, C.L., Zamble, D.B.(2013) Biochemistry 52: 1788-1801
- PubMed: 24449932
- DOI: https://doi.org/10.1021/bi301600z
- Primary Citation of Related Structures:
4IXM, 4IXN - PubMed Abstract:
GTPases are critical molecular switches involved in a wide range of biological functions. Recent phylogenetic and genomic analyses of the large, mostly uncharacterized COG0523 subfamily of GTPases revealed a link between some COG0523 proteins and metal homeostasis pathways. In this report, we detail the bioinorganic characterization of YjiA, a representative member of COG0523 subgroup 9 and the only COG0523 protein to date with high-resolution structural information. We find that YjiA is capable of binding several types of transition metals with dissociation constants in the low micromolar range and that metal binding affects both the oligomeric structure and GTPase activity of the enzyme. Using a combination of X-ray crystallography and site-directed mutagenesis, we identify, among others, a metal-binding site adjacent to the nucleotide-binding site in the GTPase domain that involves a conserved cysteine and several glutamate residues. Mutations of the coordinating residues decrease the impact of metal, suggesting that metal binding to this site is responsible for modulating the GTPase activity of the protein. These findings point toward a regulatory function for these COG0523 GTPases that is responsive to their metal-bound state.