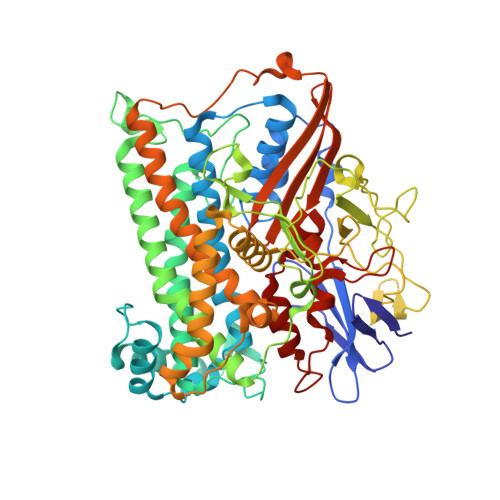
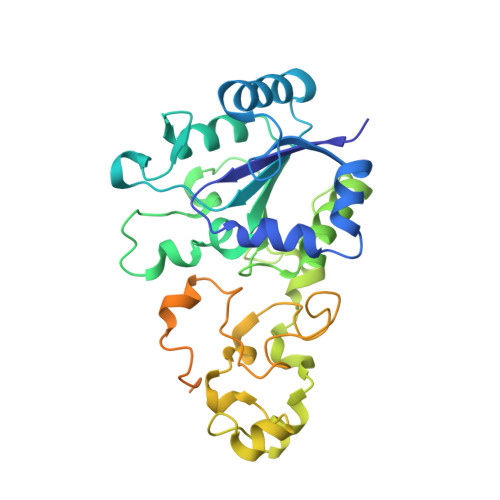
Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Frielingsdorf, S., Fritsch, J., Schmidt, A., Hammer, M., Lowenstein, J., Siebert, E., Pelmenschikov, V., Jaenicke, T., Kalms, J., Rippers, Y., Lendzian, F., Zebger, I., Teutloff, C., Kaupp, M., Bittl, R., Hildebrandt, P., Friedrich, B., Lenz, O., Scheerer, P.(2014) Nat Chem Biol 10: 378-385
- PubMed: 24705592
- DOI: https://doi.org/10.1038/nchembio.1500
- Primary Citation of Related Structures:
4IUB, 4IUC, 4IUD - PubMed Abstract:
Hydrogenases catalyze the reversible oxidation of H(2) into protons and electrons and are usually readily inactivated by O(2). However, a subgroup of the [NiFe] hydrogenases, including the membrane-bound [NiFe] hydrogenase from Ralstonia eutropha, has evolved remarkable tolerance toward O(2) that enables their host organisms to utilize H(2) as an energy source at high O(2). This feature is crucially based on a unique six cysteine-coordinated [4Fe-3S] cluster located close to the catalytic center, whose properties were investigated in this study using a multidisciplinary approach. The [4Fe-3S] cluster undergoes redox-dependent reversible transformations, namely iron swapping between a sulfide and a peptide amide N. Moreover, our investigations unraveled the redox-dependent and reversible occurence of an oxygen ligand located at a different iron. This ligand is hydrogen bonded to a conserved histidine that is essential for H(2) oxidation at high O(2). We propose that these transformations, reminiscent of those of the P-cluster of nitrogenase, enable the consecutive transfer of two electrons within a physiological potential range.
Organizational Affiliation:
1] Institut für Biologie-Mikrobiologie, Humboldt-Universität zu Berlin, Berlin, Germany. [2] Institut für Chemie, Sekr. PC14, Technische Universität Berlin, Berlin, Germany.