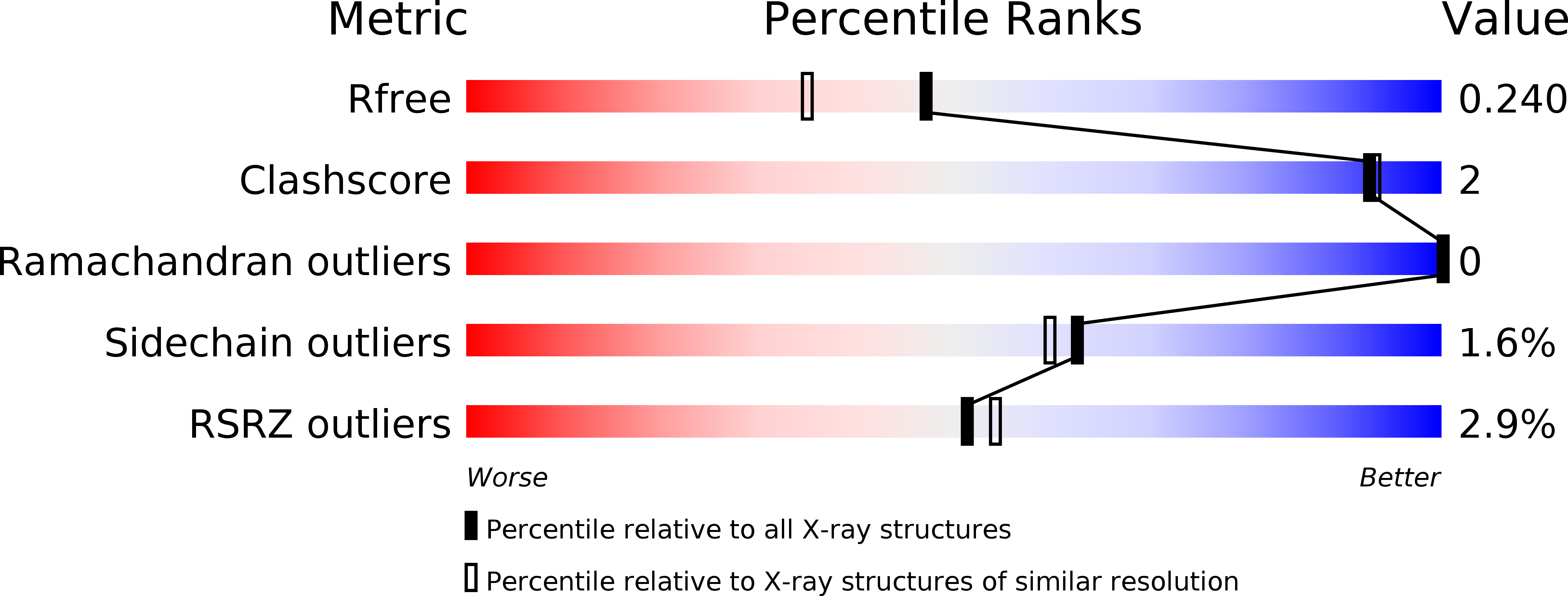
Double suppression of the G alpha protein activity by RGS proteins
Lin, C., Koval, A., Tishchenko, S., Gabdulkhakov, A., Tin, U., Solis, G.P., Katanaev, V.L.(2014) Mol Cell 53: 663-671
- PubMed: 24560274
- DOI: https://doi.org/10.1016/j.molcel.2014.01.014
- Primary Citation of Related Structures:
4IGU - PubMed Abstract:
Regulator of G protein signaling (RGS) proteins accelerate GTP hydrolysis on G protein α subunits, restricting their activity downstream from G protein-coupled receptors. Here we identify Drosophila Double hit (Dhit) as a dual RGS regulator of Gαo. In addition to the conventional GTPase-activating action, Dhit possesses the guanine nucleotide dissociation inhibitor (GDI) activity, slowing the rate of GTP uptake by Gαo; both activities are mediated by the same RGS domain. These findings are recapitulated using homologous mammalian Gαo/i proteins and RGS19. Crystal structure and mutagenesis studies provide clues into the molecular mechanism for this unprecedented GDI activity. Physiologically, we confirm this activity in Drosophila asymmetric cell divisions and HEK293T cells. We show that the oncogenic Gαo mutant found in breast cancer escapes this GDI regulation. Our studies identify Dhit and its homologs as double-action regulators, inhibiting Gαo/i proteins both through suppression of their activation and acceleration of their inactivation through the single RGS domain.
Organizational Affiliation:
Department of Pharmacology and Toxicology, University of Lausanne, Rue du Bugnon 27, 1005 Lausanne, Switzerland.