Crystal structure of the DdrB/ssDNA complex from Deinococcus radiodurans reveals a DNA binding surface involving higher-order oligomeric states.
Sugiman-Marangos, S.N., Peel, J.K., Weiss, Y.M., Ghirlando, R., Junop, M.S.(2013) Nucleic Acids Res 41: 9934-9944
- PubMed: 23975200
- DOI: https://doi.org/10.1093/nar/gkt759
- Primary Citation of Related Structures:
4HQB - PubMed Abstract:
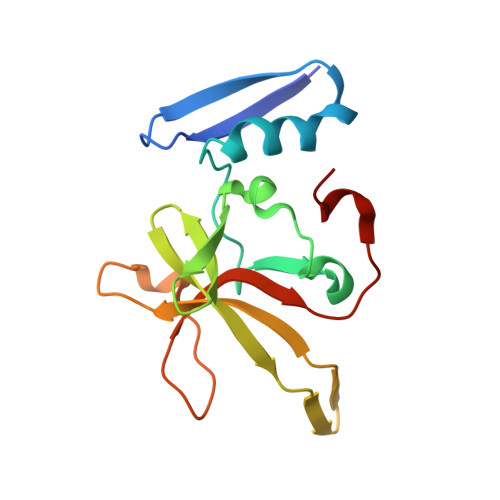
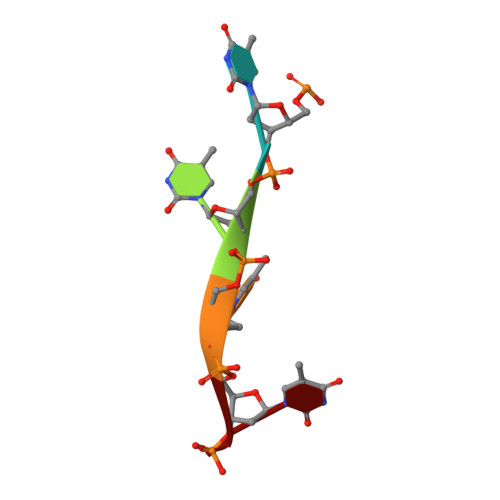
The ability of Deinococcus radiodurans to recover from extensive DNA damage is due in part to its ability to efficiently repair its genome, even following severe fragmentation by hundreds of double-strand breaks. The single-strand annealing pathway plays an important role early during the recovery process, making use of a protein, DdrB, shown to greatly stimulate ssDNA annealing. Here, we report the structure of DdrB bound to ssDNA to 2.3 Å. Pentameric DdrB was found to assemble into higher-order structures that coat ssDNA. To gain further mechanistic insight into the protein's function, a number of point mutants were generated altering both DNA binding and higher order oligomerization. This work not only identifies higher-order DdrB associations but also suggests the presence of an extended DNA binding surface running along the 'top' surface of a DdrB pentamer and continuing down between two individual subunits of the ring structure. Together this work sheds new insight into possible mechanisms for DdrB function in which higher-order assemblies of DdrB pentamers assist in the pairing of complementary ssDNA using an extended DNA binding surface.
Organizational Affiliation:
Department of Biochemistry and Biomedical Sciences and M. G. DeGroote Institute for Infectious Disease Research, McMaster University, 1200 Main Street West, Hamilton, Ontario L8N 3Z5, Canada and Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 5 Center Drive, Bethesda, MD 20892, USA.