Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release.
Luo, M., Arentson, B.W., Srivastava, D., Becker, D.F., Tanner, J.J.(2012) Biochemistry 51: 10099-10108
- PubMed: 23151026
- DOI: https://doi.org/10.1021/bi301312f
- Primary Citation of Related Structures:
4H6Q, 4H6R - PubMed Abstract:
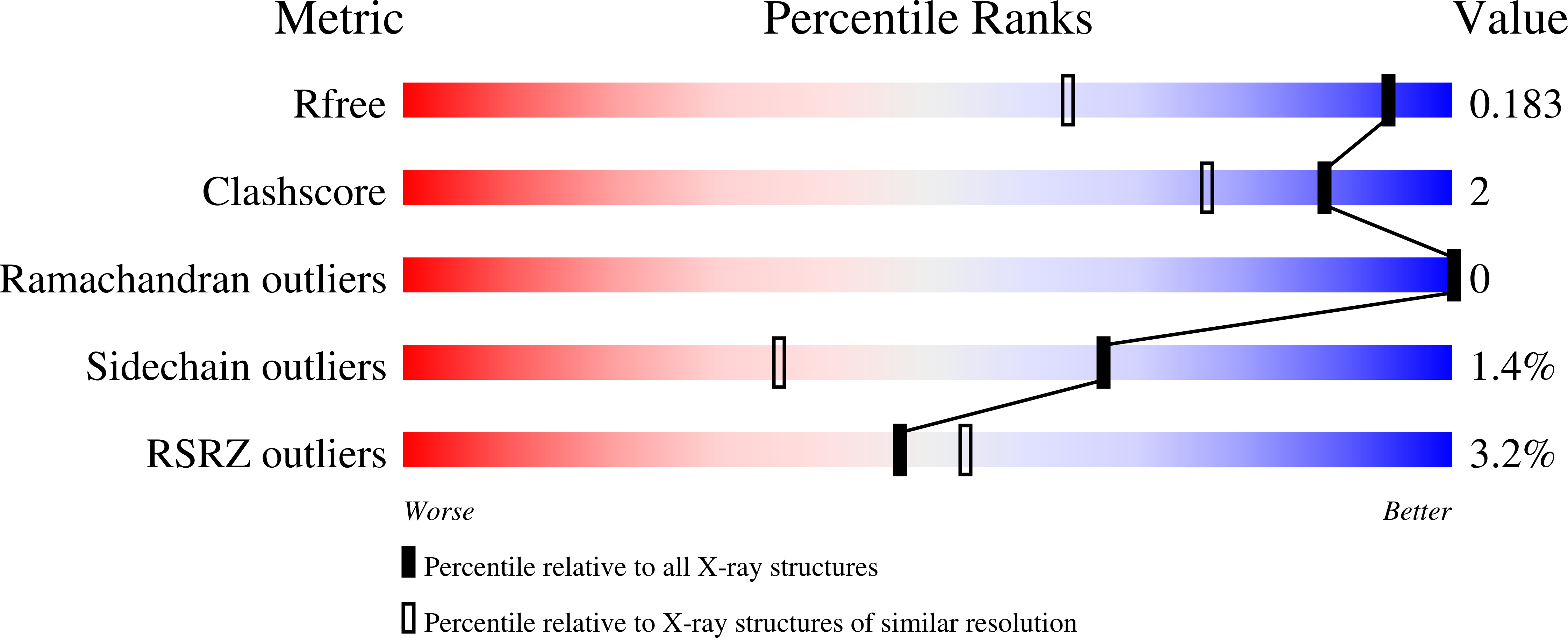
Proline dehydrogenase (PRODH) catalyzes the FAD-dependent oxidation of proline to Δ(1)-pyrroline-5-carboxylate, which is the first step of proline catabolism. Here, we report the structures of proline dehydrogenase from Deinococcus radiodurans in the oxidized state complexed with the proline analogue L-tetrahydrofuroic acid and in the reduced state with the proline site vacant. The analogue binds against the si face of the FAD isoalloxazine and is protected from bulk solvent by helix α8 and the β1-α1 loop. The FAD ribityl chain adopts two conformations in the E-S complex, which is unprecedented for flavoenzymes. One of the conformations is novel for the PRODH superfamily and may contribute to the low substrate affinity of Deinococcus PRODH. Reduction of the crystalline enzyme-inhibitor complex causes profound structural changes, including 20° butterfly bending of the isoalloxazine, crankshaft rotation of the ribityl, shifting of α8 by 1.7 Å, reconfiguration of the β1-α1 loop, and rupture of the Arg291-Glu64 ion pair. These changes dramatically open the active site to facilitate product release and allow electron acceptors access to the reduced flavin. The structures suggest that the ion pair, which is conserved in the PRODH superfamily, functions as the active site gate. Mutagenesis of Glu64 to Ala decreases the catalytic efficiency 27-fold, which demonstrates the importance of the gate. Mutation of Gly63 decreases the efficiency 140-fold, which suggests that flexibility of the β1-α1 loop is essential for optimal catalysis. The large conformational changes that are required to form the E-S complex suggest that conformational selection plays a role in substrate recognition.
Organizational Affiliation:
Department of Chemistry, University of Missouri-Columbia, Columbia, MO 65211, USA.