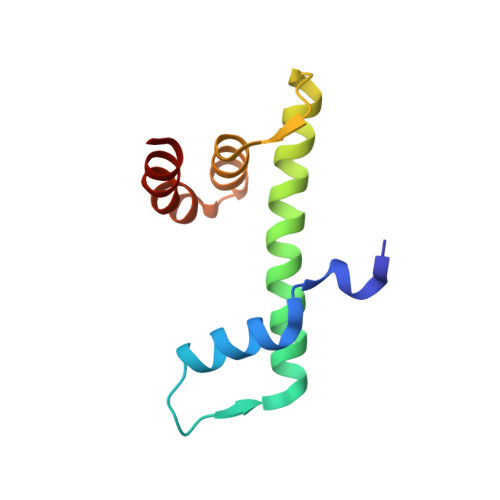
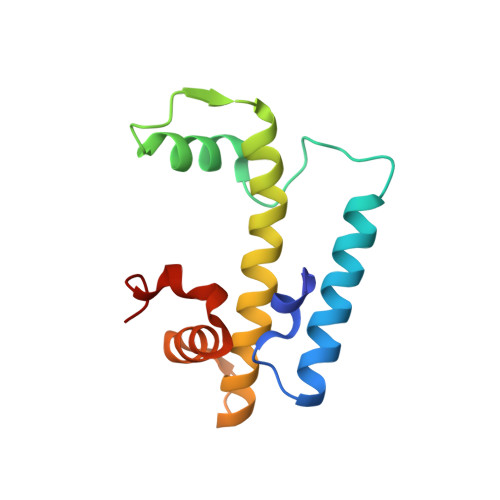
DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Huber, E.M., Scharf, D.H., Hortschansky, P., Groll, M., Brakhage, A.A.(2012) Structure 20: 1757-1768
- PubMed: 22902862
- DOI: https://doi.org/10.1016/j.str.2012.07.012
- Primary Citation of Related Structures:
4G91, 4G92 - PubMed Abstract:
The CCAAT box is a frequent element of eukaryotic promoters, and its specific recognition by the conserved heterotrimeric CCAAT-binding complex (CBC) constitutes a key step in promoter organization and regulation of transcription. Here, we report the crystal structures of the CBC from Aspergillus nidulans in the absence and in complex with double-stranded DNA at 1.8 Å resolution. The histone-like subunits HapC and HapE induce nucleosome-like DNA bending by interacting with the sugar-phosphate backbone. Minor groove sensing and widening by subunit HapB tightly anchor the CBC to the CCAAT box, as shown by structural and biochemical data. Furthermore, crucial interactions of the DNA duplex with subunit HapB provide an explanation for the sequence specificity of the CBC. The herein-described mode of transcription factor binding answers the question of how histone proteins gained sequence specificity for the CCAAT box.
Organizational Affiliation:
Center for Integrated Protein Science, Department Chemie, Lehrstuhl für Biochemie, Technische Universität München, Garching 85747, Germany.