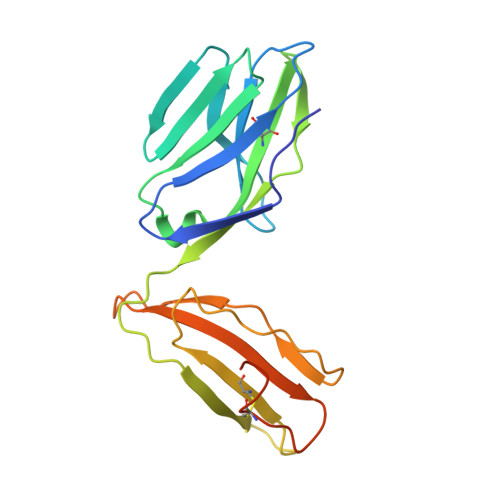
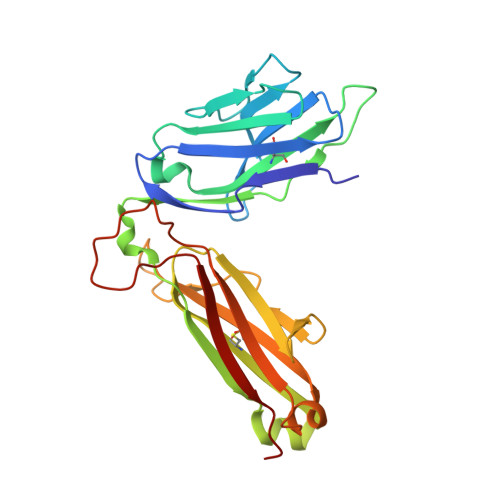
A conserved human T cell population targets mycobacterial antigens presented by CD1b.
Van Rhijn, I., Kasmar, A., de Jong, A., Gras, S., Bhati, M., Doorenspleet, M.E., de Vries, N., Godfrey, D.I., Altman, J.D., de Jager, W., Rossjohn, J., Moody, D.B.(2013) Nat Immunol 14: 706-713
- PubMed: 23727893
- DOI: https://doi.org/10.1038/ni.2630
- Primary Citation of Related Structures:
4G8E, 4G8F - PubMed Abstract:
Human T cell antigen receptors (TCRs) pair in millions of combinations to create complex and unique T cell repertoires for each person. Through the use of tetramers to analyze TCRs reactive to the antigen-presenting molecule CD1b, we detected T cells with highly stereotyped TCR α-chains present among genetically unrelated patients with tuberculosis. The germline-encoded, mycolyl lipid-reactive (GEM) TCRs had an α-chain bearing the variable (V) region TRAV1-2 rearranged to the joining (J) region TRAJ9 with few nontemplated (N)-region additions. Analysis of TCRs by high-throughput sequencing, binding and crystallography showed linkage of TCRα sequence motifs to high-affinity recognition of antigen. Thus, the CD1-reactive TCR repertoire is composed of at least two compartments: high-affinity GEM TCRs, and more-diverse TCRs with low affinity for CD1b-lipid complexes. We found high interdonor conservation of TCRs that probably resulted from selection by a nonpolymorphic antigen-presenting molecule and an immunodominant antigen.
Organizational Affiliation:
Division of Rheumatology, Immunology and Allergy, Brigham and Women's Hospital, Harvard Medical School, Boston, Massachusetts, USA. i.vanrhijn@uu.nl