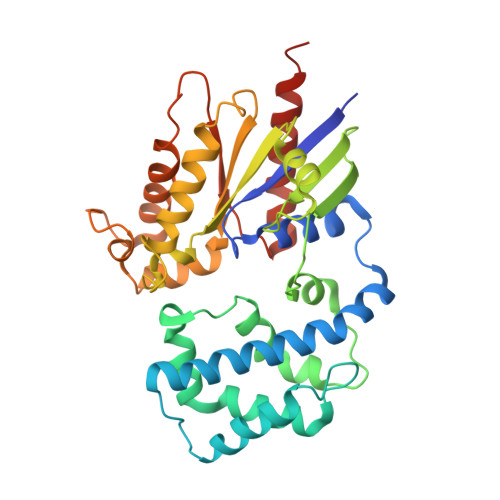
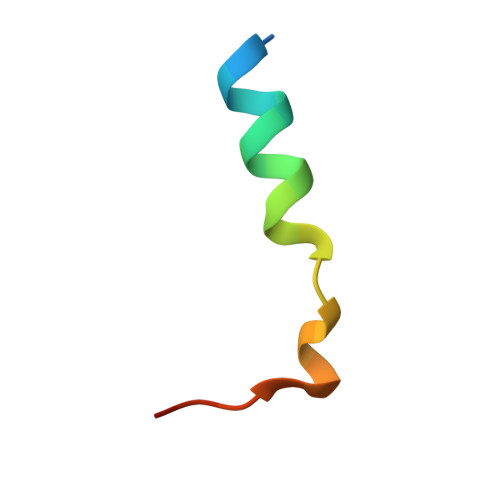
Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
Jia, M., Li, J., Zhu, J., Wen, W., Zhang, M., Wang, W.To be published.