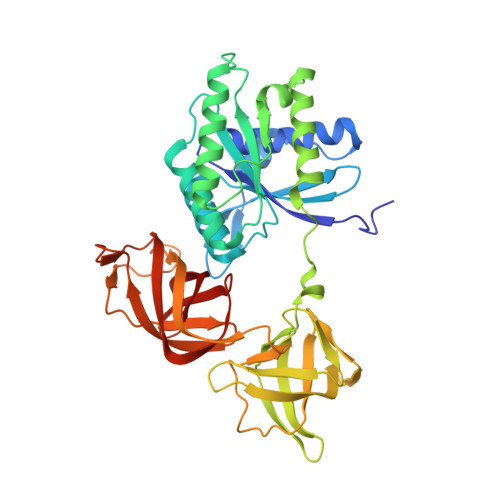
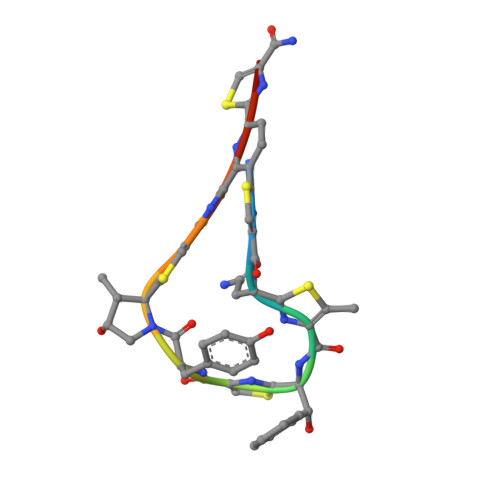
Antibiotic optimization and chemical structure stabilization of thiomuracin A.
LaMarche, M.J., Leeds, J.A., Dzink-Fox, J., Gangl, E., Krastel, P., Neckermann, G., Palestrant, D., Patane, M.A., Rann, E.M., Tiamfook, S., Yu, D.(2012) J Med Chem 55: 6934-6941
- PubMed: 22812377
- DOI: https://doi.org/10.1021/jm300783c
- Primary Citation of Related Structures:
4G5G - PubMed Abstract:
Synthetic studies of the antimicrobial secondary metabolite thiomuracin A (1) were initiated to improve chemical stability and physicochemical properties. Functional group modifications of 1 included removing the C2-C7 side chain, derivatizing the C84 epoxide region, and altering the C44 hydroxyphenylalanine motif. The resulting derivatives simplified and stabilized the chemical structure and were evaluated for antibacterial activity relative to 1. The simplified structure and improved organic solubility of the derivatives facilitated isolation yields from fermentation broths and simplified the procedures involved for the process. These advancements increased material supply for continued medicinal chemistry optimization and culminated in the identification of 2, a structurally simplified and chemically stable analogue of 1 which retained potent antibiotic activity.
Organizational Affiliation:
Global Discovery Chemistry, Novartis Institutes for Biomedical Research, Cambridge, Massachusetts 02139, United States. matthew.lamarche@novartis.com